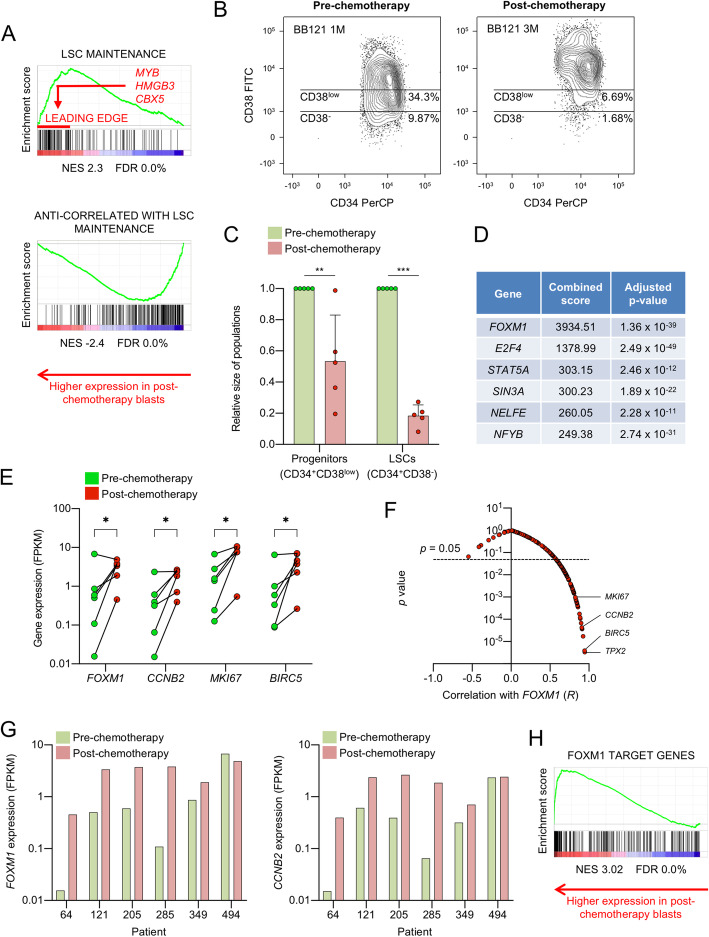
Fig. 3.
A Gene set enrichment analysis (GSEA) plots. NES, normalised enrichment score; FDR, false discovery rate. B Representative flow cytometry scatter plots show the relative size of the immunophenotypic leukemia stem and progenitor cell populations pre- and post-chemotherapy. C Mean ± SD relative size of AML stem and progenitor populations (n = 5). Sample 494 was excluded as the blasts lacked CD34 expression at presentation. ** P < 0.01, *** P < 0.001 by unpaired t-test. BB numbers indicate Biobank identifier. D Table shows enrichment of gene sets directly bound by the indicated transcription factors among the 150 genes upregulated in post-chemotherapy AML blasts. The combined score is the product of the logarithm of the adjusted p-value and the z-score (20). The analysis was performed using Enrichr (19). E Expression of the indicated genes in pre- and post-chemotherapy flow sorted AML blast populations (n = 6). FPKM, fragments per kilobase of transcript per million mapped reads. * P < 0.05 by ratio paired t-test. F Scatter plot shows correlations between absolute expression values (FPKM) for FOXM1 and the 150 genes significantly upregulated in post-chemotherapy blasts (Table S5; n = 12). R represents the Pearson product moment correlation coefficient. G Bar charts show expression of the indicated genes in all samples. FPKM, fragments per kilobase of transcript per million mapped reads. H GSEA plot shows significantly enriched expression of FOXM1 target genes in post- versus pre-chemotherapy AML blasts. NES, normalised enrichment score; FDR, false discovery rate