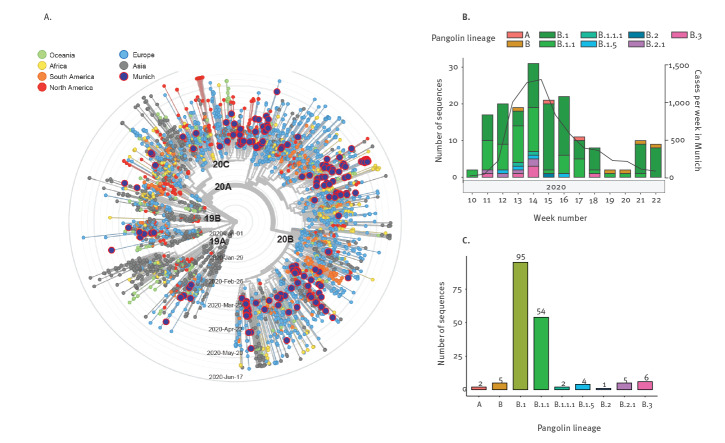
Figure 1.
Phylogenetic relationship of SARS-CoV-2 isolates from the Munich Metropolitan Region, February–May 2020 (n = 174) and global strains
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
A. Time-resolved maximum likelihood phylogeny of 3,776 global subsampled sequences available from GISAID (accession date: 6 June 2020) and 174 genomes from Munich obtained in this study (dark blue circle with red border). Nextstrain nomenclature clades are indicated above the main branches. Accession numbers are shown in Supplementary Table S3.
B. Breakdown of sequenced cases (n = 174) according to Pangolin nomenclature across calendar weeks of 2020. Total number of cases per week in Munich are plotted on the right y-axis.
C. The total number of sequenced cases is shown for each Pangolin lineage.