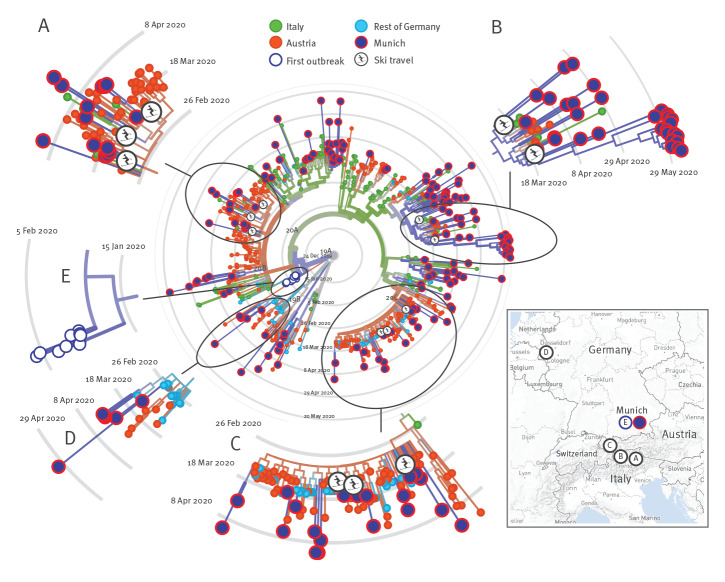
Figure 2.
Regional phylogenetic relationship of SARS-CoV-2 isolates, Munich Metropolitan Region, February–May 2020 (n = 174), with strains from Austria, Germany and Italy
SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
The 174 SARS-CoV-2 genomes obtained in this study (dark blue circles with red border) are shown in relation to subsampled sequences deposited at GISAID from Italy (green, n = 61), Austria (red, n = 88) and the rest of Germany (light blue, n = 92) in a time-resolved maximum likelihood phylogenetic tree. Accession numbers are shown in Supplementary Table S4. Sequences generated from cases with a recent travel history to skiing areas are indicated with skier symbols. Genomes of cases from the first outbreak of SARS-CoV-2 infections in the Munich area are shown as white circles with blue borders. The map indicates the geographical locations of the magnified clusters A–E.
A. Magnified insert of a cluster of sequences from cases including three travellers returning from skiing-holidays in the Dolomites, northern Italy.
B. Cluster of genomes including early cases of patients returning from South Tyrol, northern Italy.
C. Group of sequences with three ski-travellers returning from Ischgl, Tyrol, Austria.
D. Cluster of genomes with early cases from the Heinsberg area, North-Rhein-Westphalia, Germany.
E. Zoomed view of sequences from the first outbreak in the Munich area involving an introduction by a Chinese business traveller at the end of January 2020 (white circles with blue border).