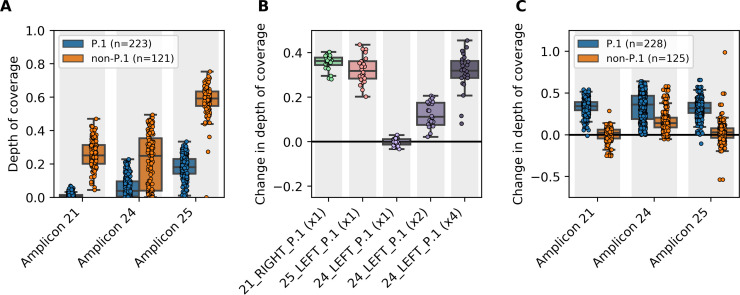
Figure 1.
(A) Predominant mutations in the P.1 lineage reduced sequencing depth across three amplicons. A total 373 clinical specimens, collected during March and April 2021 in British Columbia, Canada, were sequenced using the Freed et al. primers. Normalized mean depths of coverage are shown for three amplicons affected by mutations in primer sites (the mean depth of coverage across each library's most deeply sequenced amplicon was used for normalization). (B) Amplicon drop-out was corrected by spiking in supplemental primers. Twenty-four clinical specimens were sequenced with non-spiked and spiked primer pools, then changes in normalized mean depth of coverage were calculated for the three affected amplicons. Supplemental primers were spiked in at the same molarity as the original primers. Primer 24_LEFT_P.1 was also assayed at two-times and four-times molarity after one-times molarity did not significantly improve performance. (C) Spiked primer pools had no detrimental effects on non-P1 lineages. A total 373 clinical specimens were sequenced with non-spiked and spiked primer pools containing primers 21_RIGHT_P.1 and 25_LEFT_P.1 at one-times molarity and primer 24_LEFT_P.1 at four-times molarity. Changes in normalized mean depth of coverage were calculated for the three affected amplicons.