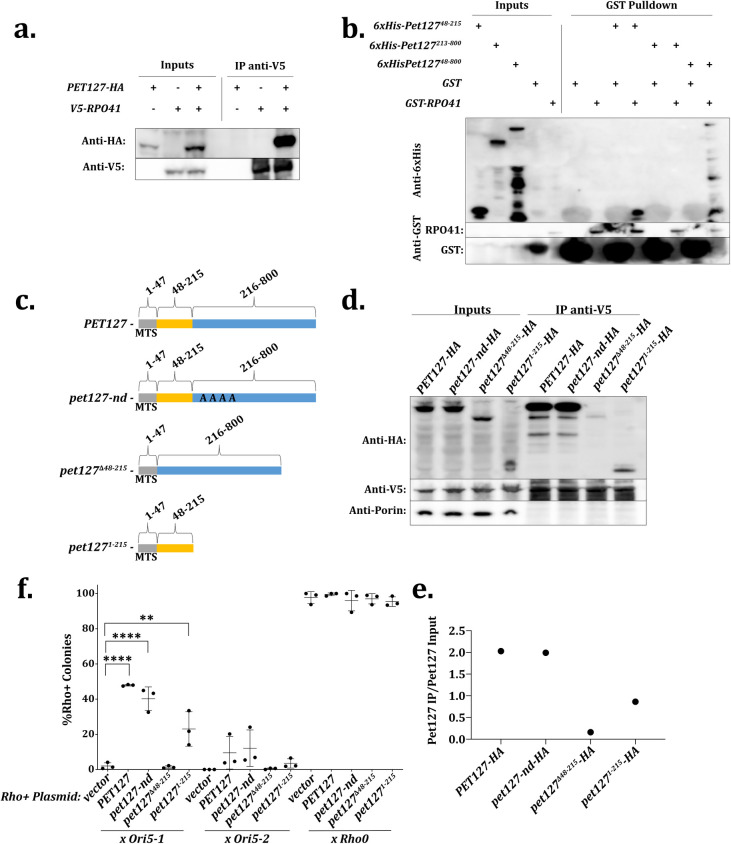
Fig 4. PET127 Binds to RPO41 and the Binding Region is Responsible for Suppression of HS Biased Inheritance.
A. Coimmunoprecipitation of Rpo41 and Pet127. Cells expressing either PET127-HA, V5-RPO41, or PET127-HA and V5-RPO41 were collected, lysed, and V5-Rpo41 was immunoprecipitated using anti-V5 conjugated beads. Samples were immunoblotted as indicated. B. Bacterial expression and lysate mixing of recombinant RPO41 and PET127 alleles. The indicated alleles were expressed in BL21 by the addition of IPTG. Cells were collected and lysed in a French press. For IPs, equal amounts of lysate from Pet127 construct expressing cells was mixed with lysate from either GST-Rpo41 or GST only expressing cells and precipitated with glutathione agarose beads. Samples were immunoblotted as indicated. C. Diagram of PET127 alleles. The predicted mitochondrial targeting sequence is amino acids 1–47 in gray [54]. Amino acids 48–215 contain the Rpo41 binding region in yellow, and amino acids 216–800 contain the region lacking Rpo41 binding activity in blue. pet127-nd is a nuclease dead allele of PET127 with amino acids predicted to be conserved active site residues changed to alanines: E346, D378, D391, and K393 [50]. D. Co-immunoprecipitation of PET127-HA alleles and V5-RPO41 as in a. Samples were immunoblotted as indicated. Anti-porin used as a negative pulldown control. E. Quantitation of IP enrichment in d. F. Quantitative mtDNA inheritance assay with rho+ high copy PET127 alleles x HS ORI5-1, HS ORI5-2, or rho0. Significance was determined using one-way ANOVA separately on each HS and rho0 cross with means compared to the vector control using Dunnett’s multiple comparison’s test (N = 3). **** indicates adjusted P-value less than 0.0001 and ** indicates an adjusted P-value of 0.0025. All other comparisons were not significantly different.