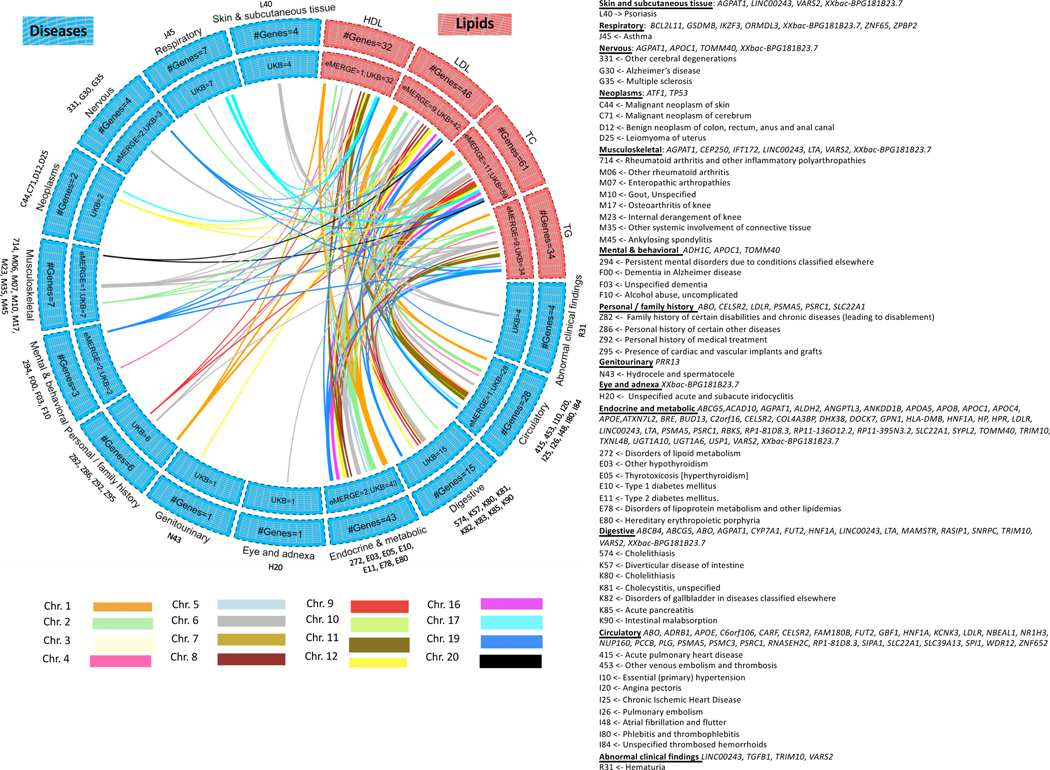
Figure 4 |. Lipid-disease pleiotropy from Xpress-PheWAS and colocalization in either eMERGE or UKB.
Circos plots indicate Bonferroni-significant genes from lipid TWAS and Xpress-PheWAS in eMERGE or UKB. We have listed the genes with no LD contamination for both lipids and diseases (i.e. coloc P[H3] < 0.5). Furthermore, these genes also have P[H4] > 0.01 from Xpress-PheWAS. Outer track, number of genes; inner track, number of genes detected in either cohort; links, genes connecting lipids (in salmon) to diseases (in blue); link thickness, number of genes; link color, chromosome. On the right-hand side, we have delineated the list of the ICD-9 or ICD-10 codes corresponding to the disease categories, along with the genes associated with each disease category.