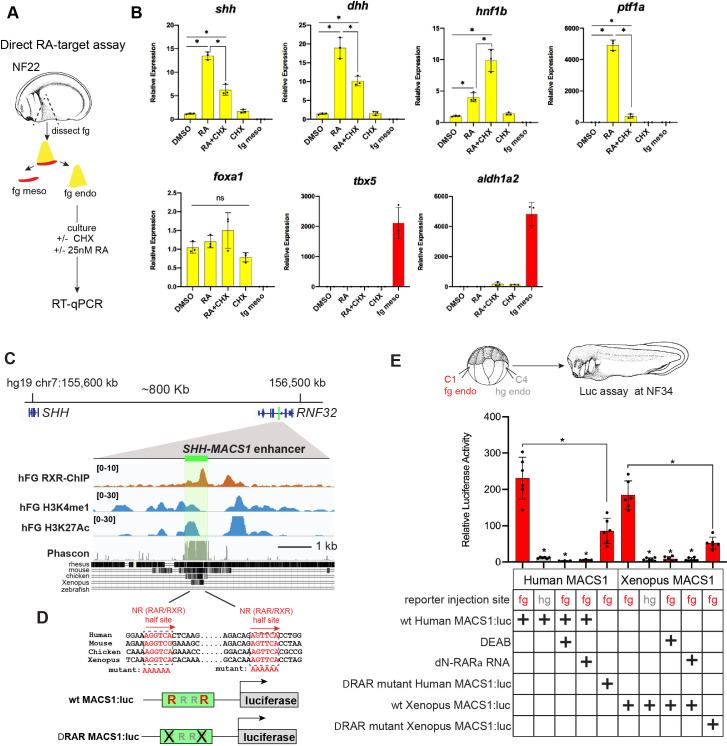
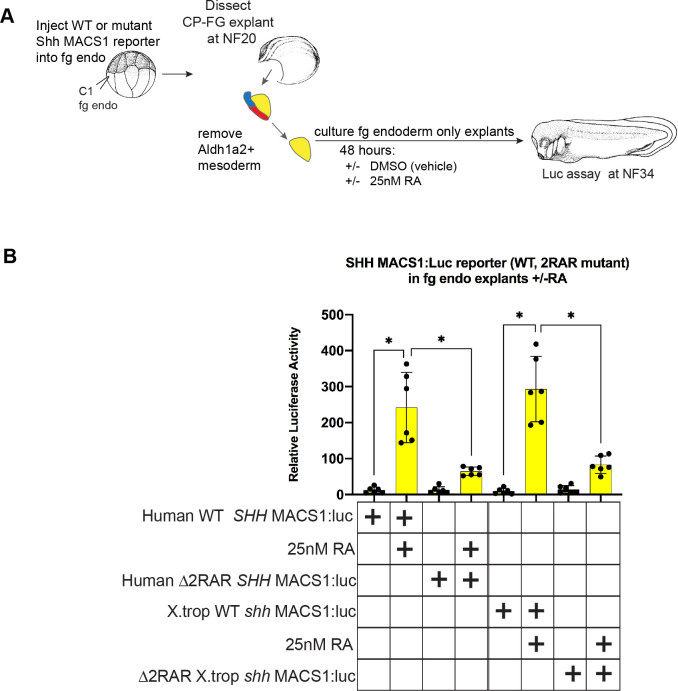
Figure 6. RA-RAR directly activates shh transcription in the Xenopus foregut endoderm via an evolutionarily conserved MACS1 enhancer.
(A) Schematic of direct RA target gene assay. Foregut endoderm (fg endo; yellow) was dissected from foregut mesoderm (fg meso; red) at NF25, pre-treated with 1 µM cycloheximide (CHX) for 2 hr prior to culture in 25 nM RA + CHX (or DMSO vehicle control) for 6 hr followed by RT-qPCR analysis. (B) RA directly activates shh and dhh expression in the presence of CHX. Graphs show mean relative expression ± standard deviation from N=3 biological replicates (four explants/replicate). Endoderm genes are shown in yellow, mesoderm makers in red confirm dissections. *p<0.05, parametric two-tailed paired t-test. (C) Genome browser of the human SHH locus showing the evolutionarily conserved MACS1 distal enhancer (green shading) embedded in an intron of the RNF32. Published ChIP-seq tracks of RXR, H3K4me1, and H3K27ac1 from hPSC-derived foregut endoderm (Vinckier et al., 2020, GSE104840; Wang et al., 2015, GSE54471). (D) MACS1 enhancer contains multiple RAR/RXR DNA-binding half sites, two of which are highly conserved. Schematics of the wild-type and mutant MACS1:luciferase reporter constructs. (E) Luciferase reporter assay in Xenopus show that the Human and X. tropicalis MACS1 enhancers are activated by RA via the RAR/RXR DNA-binding sites. 50 pg of MACS1:luciferase reporter +5 pg pRL-TK reporter were microinjected±250 pg of dominant-negative RARa RNA into either the C1 foregut (fg; red bars) or C4 hindgut (hg; gray bars) blastomeres and luciferase activity was assayed at NF34. 10 μM DEAB treatment was from NF20 to NF34. Mean relative luciferase activity ± standard deviation, from N=6 biological replicates/time point with five embryos/replicate. *p<0.05, parametric two-tailed paired t-test relative to WT MACS1:luc in the foregut (fg). Also see Figure 6—figure supplement 1, Figure 6—figure supplement 2 and related source data files. ns, not significant.