FIGURE 3.
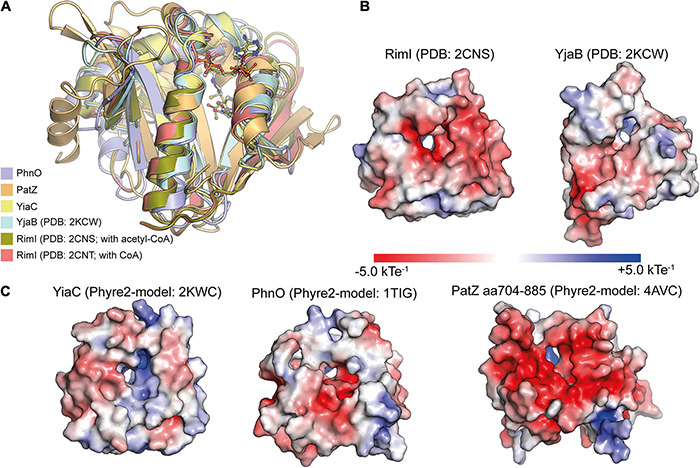
Structural characterization of E. coli GNAT protein acetyltransferases. (A) E. coli GNAT domains of protein acetyltransferases are structurally very similar. For the KATs PhnO, PatZ (aa704-885) and YiaC, Phyre2 models were created. These were superimposed with the structurally characterized KATs RimI (PDB: 2CNS and 2CNT) and YjaB (PDB: 2KCW). The catalytically important tyrosine residue suggested to act as general acid supporting resolving of the tetrahedral intermediate superimposes well. The KATs show a high degree of structural similarity showing root-mean-square-deviations (RMSD) between 0.064 and 2.319 Å toward YiaC [structural models were created with Phyre2 (Kelley et al., 2015); the figure was generated with PyMOL v.2.3.4) (Schrödinger LLC, 2000)]. (B,C) The APBS Electrostatics plugin in PyMOL was used to plot the electrostatic potential on the surfaces of the experimentally determined structures of the KATs RimI (PDB: 2CNS) and YjaB (PDB: 2KWC) (B) or the Phyre2-generated structural models of YiaC, PhnO, and PatZ (C) (Jurrus et al., 2018). All structures were oriented toward the binding site. The electrostatics on the substrate binding area differs considerably suggesting that these KATs use diverse substrates. Moreover, also the electrostatics within the active site differs suggesting that it might support catalysis to different extent. The figure was generated with PyMOL v.2.3.4 (Schrödinger LLC, 2000).
