FIGURE 4.
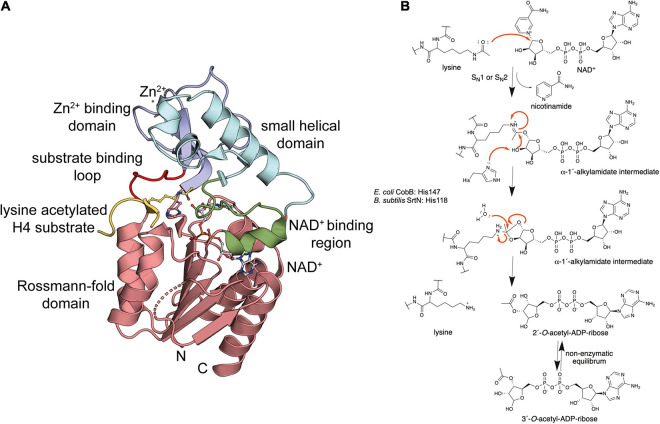
Bacterial sirtuins are NAD+-dependent lysine-deac(et)ylases. (A) Structure of the E. coli sirtuin deacetylase CobB in complex with an lysine acetylated human histone 4 substrate (PDB: 1S5P). The CobB structure was superimposed to a structure of human SIRT2 in complex with NAD+ (PDB: 4RMG) to show localization of NAD+. CobB contains a Rossmann-fold domain (salmon) composed of a six-stranded parallel β-sheet flanked by several α-helices containing the NAD+-binding site. An NAD+-binding region (green) contributing to NAD+-binding connecting Rossmann-fold and small helical domain (light blue). The Zn2+-binding domain (dark blue) contains a structural Zn2+-ion that is coordinated by two pairs of conserved Cys-residues (only two are visible in this structure). The substrate binding loop (red) connects the Zn2+-binding domain and the Rossmann-fold domain [the figure was generated with PyMOL v.2.3.4 (Schrödinger LLC, 2000)]. (B) Catalytic mechanism exerted by sirtuins. Sirtuins are NAD+-dependent lysine deac(et)ylases enzymes that use NAD+ as stoichiometric co-substrate for catalysis. Initially, the carbonyl-oxygen of the lysine’s ac(et)yl-group as a nucleophile performs an attack of the electrophilic C-1′ of the NAD+ ribose. This results in fast release of nicotinamide subsequent formation of a C-1′-O-alkylamidate intermediate. Several steps are needed to resolve this intermediate. These include a hydrolysis step, as shown, resulting in formation of the deac(et)ylated lysine and 2′-O-acetyl-ADP-ribose, which exists in a non-enzymatic equilibrium with 3′-O-acetyl-ADP ribose [figure redrawn and modified from Smith and Denu (2006), Ali et al. (2018), Teixeira et al. (2020), and Blasl et al. (2021)].