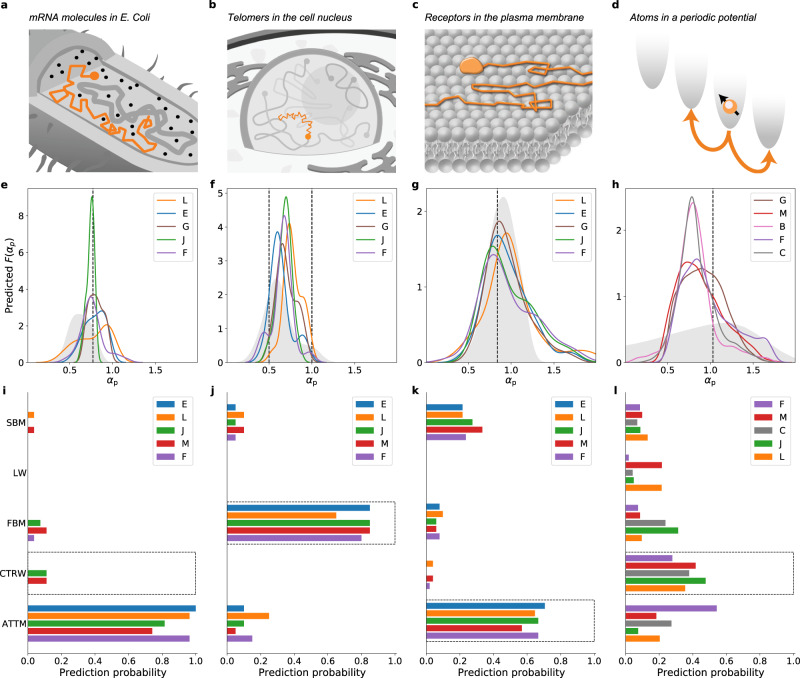
Fig. 5. Analysis of experimental datasets.
a–d, Schematic representation of the experiments analyzed in the contest of the AnDi challenge: 2D motion of mRNA molecules inside live E. coli cells (47, a); 2D motion of telomeres in the nucleus of mammalian cells (38,48, b); 2D motion of biomolecular receptors moving on the membrane of mammalian cells (20, c); and 1D motion of single atoms moving in a 1D periodic optical potential (49, d). e–h Histograms of the estimation of the anomalous diffusion exponent αp predicted by top teams for trajectories from experimental datasets. Gray areas correspond to the results of the baseline method TA-MSD. Dashed lines indicate the original estimations of α provided by Refs. 47 (e),38,48 (f),20 (g), and49 (h). i–l Histograms of the diffusion model predicted by top teams for trajectories from experimental datasets. Dashed boxes indicate the original classifications provided by Refs. 47 (i),38,48 (j),20 (k), and49 (l). We show predictions obtained by the top 5 teams for the corresponding subtask. For the last dataset, we further selected the teams based on their performance on short (L ≈ 10) trajectories. All results for the analysis of the experimental data are presented in Supplementary Figs. 21-28.