Figure. The application and findings of single-cell RNA sequencing (scRNA-seq ) analysis in aortic aneurysms.
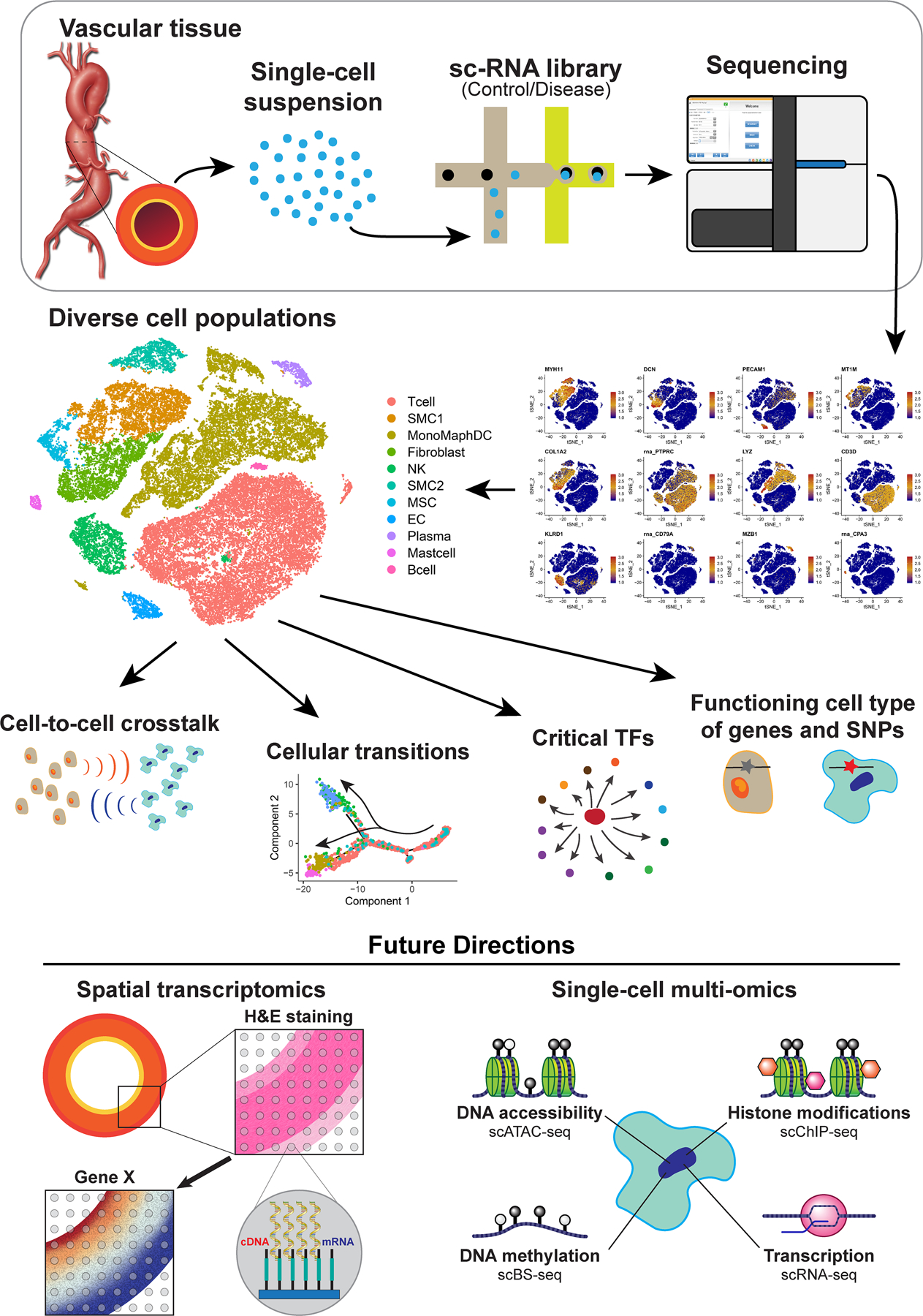
ScRNA-seq starts with the processing of aortic tissue into a single-cell suspension that is submitted for library preparation (the 10X Genomics platform is shown as an example) and then sequencing. ScRNA-seq analysis has revealed the cellular heterogeneity, cellular transitions, communications among cell populations, and critical transcription factors in the aortic wall. ScRNA-seq has also contributed to our understanding of the pathogenesis of aortic aneurysms through the identification of the functioning cell type of aneurysm-related genes and genetic variants. Future directions of scRNA-seq applications in studies of aortic aneurysms include the integration of scRNA-seq with spatial transcriptomics and single-cell multiomics. ScATAC-seq, single-cell sequencing assay for transposase-accessible chromatin; scBS-seq, single-cell bisulfite sequencing; scChIP-seq, single-cell chromatin immunoprecipitation sequencing; SNPs, single-nucleotide polymorphisms; TFs, transcription factors.
