FIGURE 3.
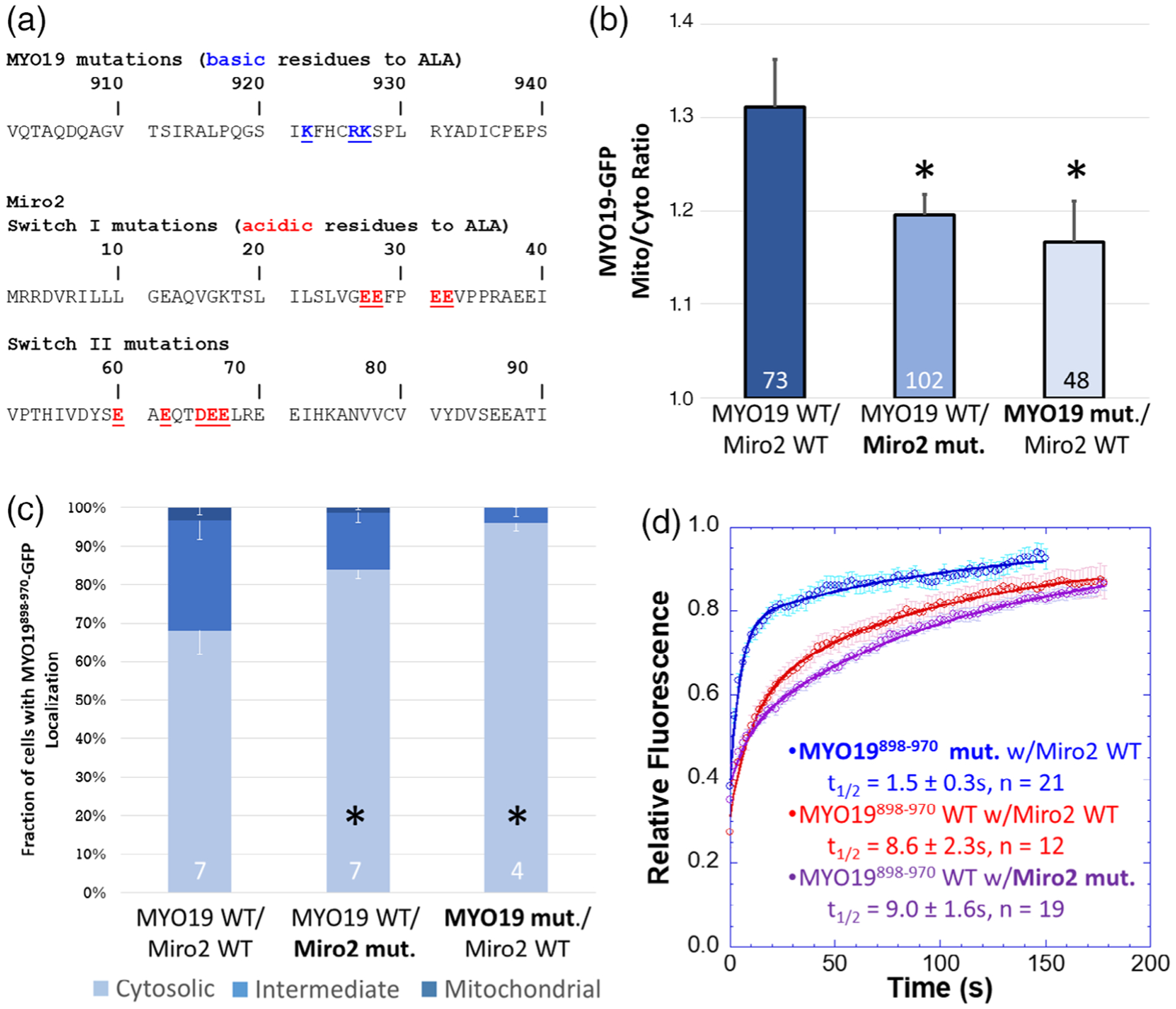
A subset of well-conserved, charged residues are essential for Miro2-dependent localization of MYO19898–970. (a) Clustal Omega multiple sequence alignments of the MYO19 MyMOMA domain and the Miro2 N-terminal GTPase domain identified well-conserved, charged residues in each domain, which were mutated to alanine to determine their contribution to the MYO19/Miro2 interaction. (b) Mutating either the basic residues in MYO19898–970-GFP to alanine or the acidic residues in mchr-Miro2 to alanine decreased the localization of MYO19898–970-GFP to mitochondria, as determined by the mito/cyto ratio (*p < 0.05, Dunnett’s test vs. MYO19898–970-GFP wild type coexpressed with mchr-Miro2 wild type). (c) Mutations similarly decreased the fraction of cells displaying a cytosolic staining pattern for GFP, whether those mutations were to MYO19 or Miro2 (*p < 0.05, Dunnett’s test vs. MYO19898–970-GFP wild type coexpressed with mchr-Miro2 wild type). (d) FRAP analysis of MYO19898–970-GFP mutant shows faster exchange kinetics than wild type, consistent with basic residues in the MYO19 mediating interactions with Miro2. The exchange kinetics for wild type MYO19898–970-GFP with the mutant mchr-Miro2 construct were similar to exchange kinetics with the wild type mchr-Miro2 wild type construct. This is likely due to endogenous Miro recruiting MYO19898–970-GFP. Multiple sequence alignments can be found in the Figure S4. Numbers at the base of the bars indicate the number of replicates. Error bars represent SEM. FRAP kinetic parameters are listed in Table 1
