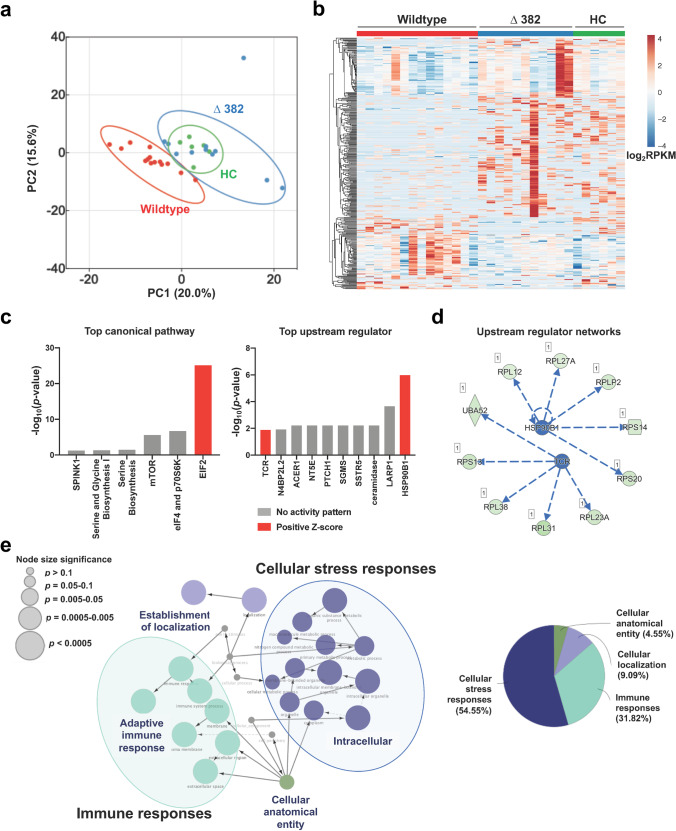
Fig. 2.
Effects of 382-nt deletion in SARS-CoV-2 ORF8 genome (Δ382) on whole blood transcriptome of COVID-19 patients. RNA-seq of whole blood from COVID-19 patients infected with WT (n = 14) and Δ382 SARS-CoV-2 (n = 11) at the acute phase of infection (SARS-CoV-2 PCR-positive; median 8 days PIO) was performed. Only samples with RNA integrity number > 6 were sent for sequencing and included in the analysis a PCA of COVID-19 patients and healthy controls based on DEGs, with p-value < 0.01 and |FC|> 2. b Heatmap of 358 DEGs, scaled based on log2RPKM values, with blue and red colors indicating low and high expressions, respectively. c Top canonical pathways and upstream regulators identified by IPA based on the DEGs. Bar graphs are ranked by significance, with red indicating positive predicted activation Z-scores and gray indicating undetermined directionality. d An integrated network of HSP90B1 and TCR and their targeted genes. Stimulation of HSP90B1 and TCR leads to overexpression of the downstream genes. e GO pathway term enrichment networks of DEGs using Cytoscape add-on ClueGO. Each of the GO terms is statistically significant (Benjamini–Hochberg correction < 0.05). The filled colored circles (nodes) represent a statistically significant enriched parent GO term. The lines (edges) between nodes show overlapping genes within terms, with node size representing the term enrichment significance. The overview chart shows the distribution of the functionally grouped GO terms. The cut-off for terms in the functionally grouped networks was set at p-value < 0.05. WT, wildtype; PCA, principal component analysis; DEGs, differentially expressed genes; FC, fold change; RPKM, reads per kilobase per million reads mapped; PCR, polymerase chain reaction; IPA, Ingenuity Pathway Analysis; HSP90B1, heat shock protein 90 kDa beta member 1; TCR, T cell receptor; GO, gene ontology