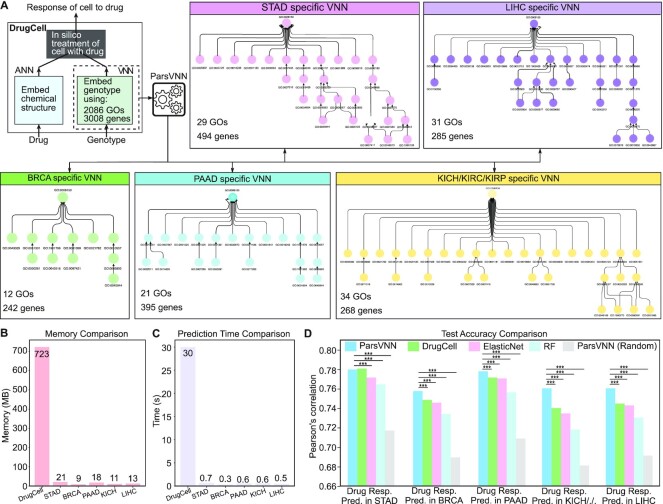
Figure 3.
The architectures of five cancer-specific ParsVNN models and their performance on test samples. (A) The architecture of DrugCell. The resulting architectures of ParsVNN models for STAD, BRCA, PAAD, KICH/KIRC/KIPR and LIHC cancer types after applying ParsVNN to the original DrugCell model using cancer-specific training samples. (B) Memory comparison of DrugCell and the pruned models. (C) Prediction time comparison of DrugCell and the pruned models. Prediction time is the average forward passing time of the deep learning models for 10 000 inputs over 10 times. (C) Performance comparison between ParsVNN and other competing methods on drug response prediction for five different cancer types. RF stands for random forest. We use the Pearson’s correlation between the predicted drug responses and the observed drug responses as the evaluation criterion. The asterisks (***) indicate the P-value <0.0001.