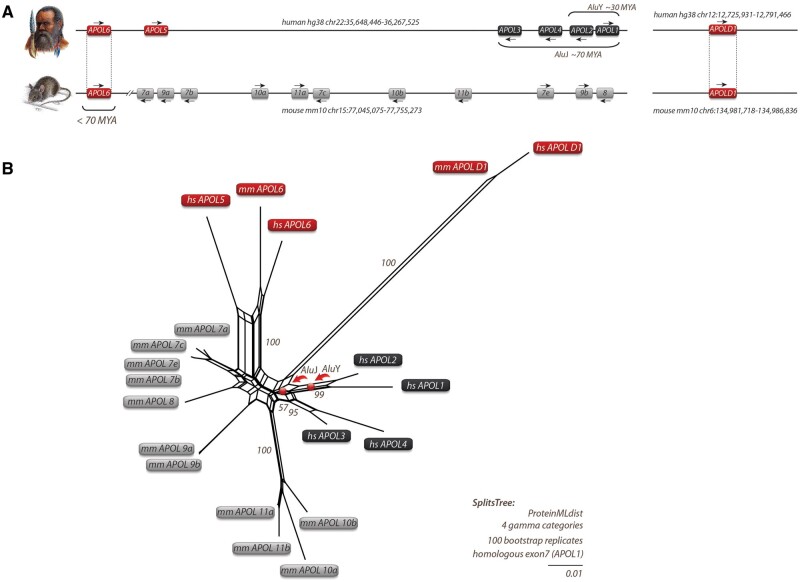
Fig. 1.
Local APOL gene family and SplitsTree reconstructions for humans and mice. (A) Genomic location of APOL genes from human and mouse with genomic coordinates. Arrows indicate gene orientations. APOLD1 evolved early in vertebrates about 300 Ma, whereas APOL6 was probably inherited by a human–mouse ancestor that lived about 70 Ma. (B) SplitsTree reconstruction of APOL genes based on protein sequences. A similar tree topology was derived by maximum likelihood and Bayesian tree reconstructions (supplementary data SD1, Supplementary Material online). The APOL genes of mice (mm for Mus musculus) are indicated in gray boxes. Black boxes represent human APOL genes (hs for Homo sapiens). The central parallelograms of the reconstruction represent conflicting phylogenetic signals. The tree reconstruction reveals a common origin of mm and hs APOL6 (red boxes). Bootstrap values are shown for representative branches. Balls indicate the clade-supporting integrations of AluJ and AluY elements. The human APOL2 and APOL1 genes share an orthologous AluY element. The orthologous AluY transposons were already present in Catharrine primates (diverged about 30 Ma). The diagnostic AluJ elements were detected at orthologous positions in APOL1-4 in all investigated primates and were probably inherited from a common primate ancestor of about 70 Ma. The double-slash in the mouse locus indicates the exclusion of a large genomic region. Human and mouse APOLD1 are located on chromosomes 12 and 6, respectively, and diverged significantly from other APOL genes.