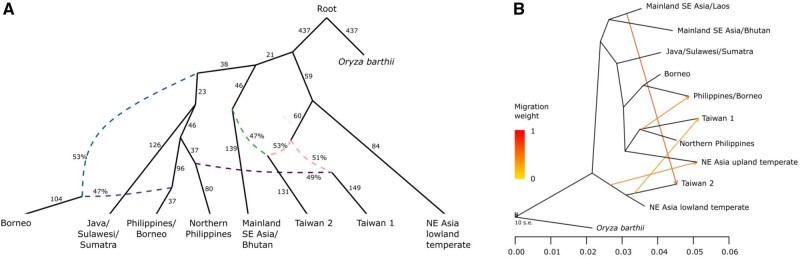
Fig. 3.
Admixture graph and TREEMIX models of japonica rice. (A) Admixture graph Kd = 9 japonica subpopulations (maximum |z-score| = 2.907), rooted with Oryza barthii as an outgroup. This graph represents topology that is consistent between models for all lower values of K. Solid lines with arrowheads indicate lineages with uniform genetic ancestries, with the scaled drift parameter f2 shown next to these lines. Dashed lines lead to subpopulations with mixed ancestries, with the estimated proportion of ancestry indicated by the percentage values. (B) Maximum likelihood trees based on Treemix. When a subpopulation has multiple ancestry sources, it forms a clade with one of the sources whereas an accompanying arrow indicates shared ancestry with the other source; this analysis is suggestive (but not conclusive) on the level and direction of gene flow.