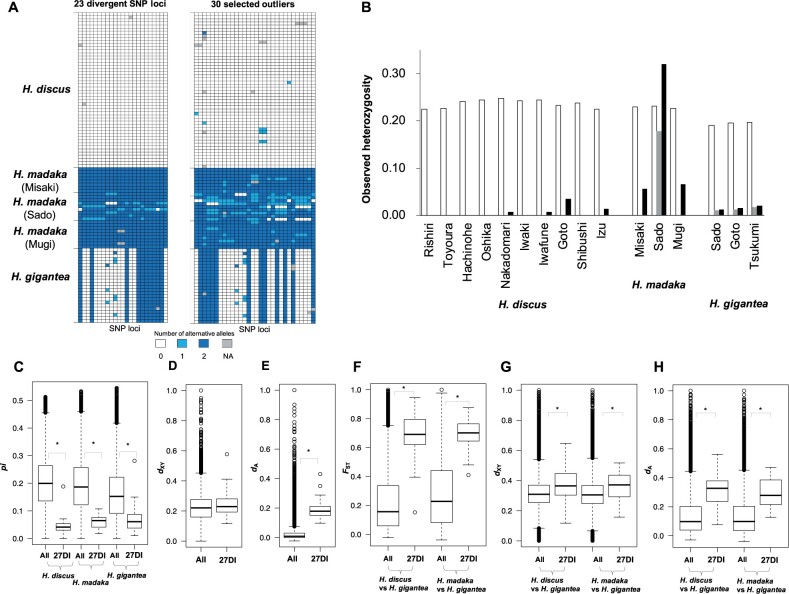
Fig. 5.
(A) Genotypes in 23 divergent and 30 selected outlier SNP loci showing the genetic divergence between Haliotis discus and H. madaka abalones. NA shows loci that were not gentyped. (B) Observed heterozygosity in each of the three Western Pacific abalone populations across all SNP loci (white), 23 divergent SNP loci (gray), and 30 selected outlier (black). (C) Boxplot of nucleotide diversity (pi) in H. discus, H. madaka, and H. gigantea in all and 27 differentiation islands (27DI). (D) Boxplot of absolute sequence divergence (dXY) between H. discus and H. madaka in all and 27DI. (E) Boxplot of net sequence divergence (dA) between H. discus and H. madaka in all and 27DI. (F) Boxplot of sliding-window FST values between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. (G) Boxplot of dXY between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. (H) Boxplot of dA between H. discus and H. gigantea, and between H. discus and H. gigantea in all and 27DI. Asterisks show significant differences (Wilcoxon rank-sum test; P < 0.05).