Abstract
Insects detect odors using an array of odorant receptors (ORs), which may expand through gene duplication. How and which new functions may evolve among related ORs within a species remain poorly investigated. We addressed this question by functionally characterizing ORs from the Eurasian spruce bark beetle Ips typographus, in which physiological and behavioral responses to pheromones, volatiles from host and nonhost trees, and fungal symbionts are well described. In contrast, knowledge of OR function is restricted to two receptors detecting the pheromone compounds (S)-(–)-ipsenol (ItypOR46) and (R)-(–)-ipsdienol (ItypOR49). These receptors belong to an Ips-specific OR-lineage comprising seven ItypORs. To gain insight into the functional evolution of related ORs, we characterized the five remaining ORs in this clade using Xenopus oocytes. Two receptors responded primarily to the host tree monoterpenes (+)-3-carene (ItypOR25) and p-cymene (ItypOR27). Two receptors responded to oxygenated monoterpenoids produced in larger relative amounts by the beetle-associated fungi, with ItypOR23 specific for (+)-trans-(1R, 4S)-4-thujanol, and ItypOR29 responding to (+)-isopinocamphone and similar ketones. ItypOR28 responded to the pheromone E-myrcenol from the competitor Ips duplicatus. Overall, the OR responses match well with those of previously characterized olfactory sensory neuron classes except that neurons detecting E-myrcenol have not been identified. The characterized ORs are under strong purifying selection and demonstrate a shared functional property in that they all primarily respond to monoterpenoids. The variation in functional groups among OR ligands and their diverse ecological origins suggest that neofunctionalization has occurred early in the evolution of this OR-lineage following gene duplication.
Keywords: functional characterization, neofunctionalization, odorant receptor, olfaction, purifying selection, Xenopus oocyte
Introduction
Olfaction is of utmost importance to the ecology of insects, with odors mediating fitness-related activities such as mate choice and the search for food and oviposition sites, and maintenance of symbioses with microbes (Hansson and Stensmyr 2011; Andersson et al. 2015; Fleischer et al. 2018; Kandasamy et al. 2019). Odors are detected by an expansive gene family encoding odorant receptors (ORs), which are expressed in the olfactory sensory neurons (OSNs) in the insect antennae (Clyne et al. 1999). As such, the ORs are crucial to insect life and they underlie the diverse chemical ecologies, adaptations, niche specializations, and evolutionary divergence seen in the Insecta class, which dominates most habitats of our planet. Yet, the functional evolution of this gene family is poorly understood (but see, e.g., Ramdya and Benton 2010; de Fouchier et al. 2017; Yuvaraj et al. 2017; Guo et al. 2021), and little is known with regards to how new functions evolve in these receptors, especially in non-model insects. This applies to both the molecular mechanics that underlie specificity changes (but see Leary et al. 2012; Hopf et al. 2015) and the ecological selection pressures that drive the evolution of the ORs.
New OR genes originate through gene duplication. Hence, the duplicated receptors that are retained and expressed will initially have the same odor specificity as their parental ORs, but may later acquire neutral or adaptive mutations that alter their specificity (Nei et al. 2008; Andersson et al. 2015). One may therefore predict that duplicated “offspring” OR paralogues are likely to adopt similar functions as their “parent” ORs, that is, detecting compounds with shared chemical characteristics (but see also Adipietro et al. 2012). In this study, we investigated the functional evolution of OR paralogues within a clade that houses seven receptors from the Eurasian spruce bark beetle Ips typographus L. (Coleoptera; Curculionidae; Scolytinae).
As keystone species in forest ecosystems, bark beetles play an important role in the decomposition of wood and recycling of nutrients (Edmonds and Eglitis 1989; Raffa et al. 2016). However, a minority of bark beetle species are able to kill healthy trees during outbreak periods, which has become an increasing threat to conifer forests, causing great economic loss (Raffa 2001; Kausrud et al. 2011; Raffa et al. 2016; Biedermann et al. 2019). In Eurasia, I. typographus is considered the most serious pest of Norway spruce (Picea abies) and it also attacks other species of spruce across its range (Økland and Bjørnstad 2003). When I. typographus population density surpasses a critical threshold, the defense of healthy trees can be overcome through mass-attacks, and entire forest landscapes can be quickly transformed (Wyatt 2014; Raffa et al. 2016). The mass-attacks are coordinated by a male-produced aggregation pheromone consisting of (4S)-cis-verbenol and 2-methyl-3-buten-2-ol, attracting both sexes (Schlyter et al. 1987). The attraction is, however, inhibited by several other pheromone compounds produced by I. typographus during the later attack phases or by heterospecific beetles, including E-myrcenol and specific enantiomers of verbenone, ipsenol, and ipsdienol (Francke et al. 1980: Birgersson et al. 1984; Schlyter et al. 1989, 1992).
In addition to the beetle-produced semiochemicals, the pheromone attraction is modulated by volatiles from nonhost trees and defense compounds from the host (Zhang and Schlyter 2004; Andersson et al. 2010; Schiebe et al. 2012; Binyameen et al. 2014; Unelius et al. 2014). Recent laboratory studies have also demonstrated the importance of volatiles in the maintenance of symbioses between I. typographus and its fungal associates, with beetle preferences for certain fungi being mediated via olfactory cues (Kandasamy et al. 2019). Once inoculated in a tree, these fungi may provide nutrients to maturing beetles by direct feeding (Kandasamy et al. 2019), metabolize host tree defenses (Kandasamy et al. 2021), and possibly accelerate tree death (Horntvedt et al. 1983). Extensive efforts have been made to understand the ecological roles of I. typographus-associated compounds and to characterize the OSNs that specifically detect them. To date, a total of 23 strongly responding OSN classes have been reported. The key ligands of these neurons include pheromone compounds from conspecific and heterospecific bark beetles, volatiles from host and nonhost plants, and compounds produced by fungal symbionts (Mustaparta et al. 1984; Tømmerås et al. 1984; Tømmerås 1985; Andersson et al. 2009; Andersson 2012; Raffa et al. 2016; Kandasamy et al. 2019; Schiebe et al. 2019; Zhao et al. 2019).
The odor selectivity of an OSN depends on the characteristics of the OR that is expressed in that neuron. Hence, functional characterization of ORs is important for understanding the evolution of olfactory specialization. Despite being the largest insect order, functional information of ORs from Coleoptera is, however, limited compared with moths (e.g., Grosse-Wilde et al. 2007; Zhang and Löfstedt 2015; Zhang et al. 2016; de Fouchier et al. 2017; Guo et al. 2021), flies (e.g., Hallem and Carlson 2006; Mansourian and Stensmyr 2015), and mosquitos (Carey et al. 2010; Wang et al. 2010). To our knowledge, only seven beetle ORs have been characterized so far, of which two belong to I. typographus (Mitchell et al. 2012; Wang et al. 2020; Antony et al. 2021: Mitchell and Andersson 2020; Yuvaraj et al. 2021). In our previous study, the two I. typographus ORs (ItypOR46 and ItypOR49), responded specifically to the pheromone compounds (S)-(−)-ipsenol and (R)-(−)-ipsdienol, respectively. These ORs are part of an Ips-specific (based on currently available ORs from bark beetles) OR-lineage radiation comprising seven ItypORs (Yuvaraj et al. 2021). This radiation is present within the coleopteran OR subfamily named Group 7, which is highly expanded in the Curculionidae family (Andersson et al. 2013, 2019; Mitchell et al. 2020). The other five ORs (ItypOR23, OR25, OR27, OR28, and OR29) in this clade did not respond to any stimulus when tested in HEK293 cells, which was likely due to insufficient protein levels (Yuvaraj et al. 2021). Because this clade represents the largest highly supported OR-radiation in the antennal transcriptome of this species and many of the OR genes are highly expressed (Yuvaraj et al. 2021), we hypothesize that these ORs have key ecological functions. Thus, targeting the five remaining ORs using a different expression system could provide information on the functional evolution of ORs in this species and also shed light on the general question of which ecological functions may evolve among OR paralogues within species- or genus-specific radiations. Hence, the primary aim of this study was to functionally characterize these five ORs using Xenopus oocytes and a large panel of ecologically relevant compounds. We asked whether the clade houses additional ORs specifically detecting other bark beetle pheromone compounds, or whether the ORs detect structurally similar chemicals (i.e., oxygenated monoterpenoids) of different biological origins and ecological meanings. The second aim was to link the responses of the functionally characterized ItypORs with the responses of previously identified OSN classes.
Results
Characteristics and Sequence Evolution of the ORs in the Ips-Specific Clade
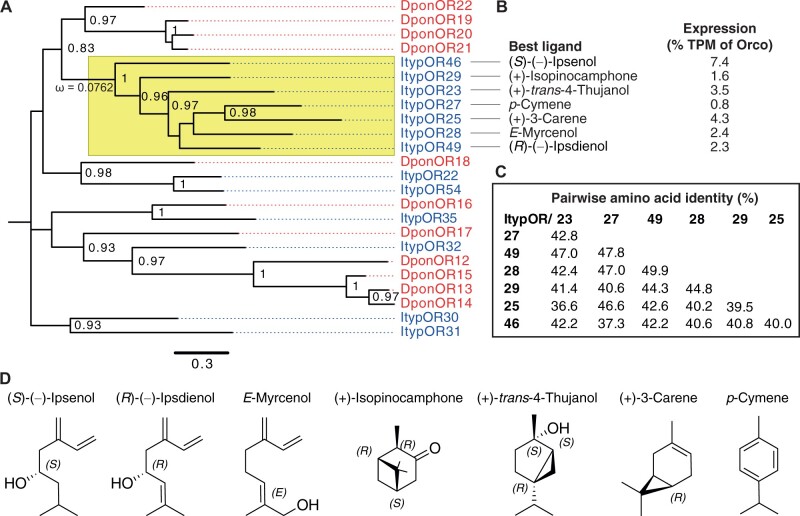
The five ItypORs targeted for functional characterization are part of an Ips-specific OR-lineage comprising seven ItypORs, and this lineage belongs to the monophyletic OR Group 7 in beetles (Mitchell et al. 2020; Yuvaraj et al. 2021). The phylogenetic relationships of these ORs and a subset of additional Group 7 ORs from I. typographus and the mountain pine beetle Dendroctonus ponderosae (“Dpon”) are shown in figure 1A. The expression levels (relative to Orco; data from Yuvaraj et al. 2021) and proposed key ligands of the seven ItypORs (based on our oocyte recordings; see below) are shown in figure 1B and D. Except for ItypOR27, these OR genes are all among the top 20 (out of 73) most highly expressed OR genes in the antennae of this species (Yuvaraj et al. 2021). The pair-wise amino acid identity amongst the seven ORs ranges from 36.6% to 49.9% (fig. 1C).
Fig. 1.
(A) Unrooted maximum-likelihood tree showing the relationships among select Group 7 (Mitchell et al. 2020) ORs from Ips typographus (“Ityp”; blue) and Dendroctonus ponderosae (“Dpon”; red). The tree is based on a MAFFT alignment of amino acid sequences and constructed using FastTree 2.1.11. The clade containing the seven ItypORs is highlighted in yellow and the strong purifying selection (ω = 0.0762) is indicated on the branch. Numbers at nodes are local support values, calculated using the SH test implemented in FastTree (SH values below 0.7 are not shown). (B) The primary ligands and expression levels (transcripts per million, TPM) relative to Orco of the seven ItypORs (data from Yuvaraj et al. 2021). (C) The pair-wise amino acid identities amongst the seven ORs, based on a MAFFT alignment. (D) Chemical structures of the main ligand for each of the seven ItypORs in the clade.
We evaluated the selective pressure acting on the seven ItypORs by calculating the ratio of nonsynonymous to synonymous substitutions (dN/dS or ω) using PAML (Yang 1997; Yang and Nielsen 1998). We first tested whether the clade has a uniform dN/dS ratio (one ratio model, M0) or variable ratios on different branches (free ratio model, M1) by likelihood ratio test (LRT). The result showed that the one ratio model could not be rejected (P = 0.19), and the dN/dS is 0.07623 (≪1) for this clade. We next examined whether there are positively selected sites across the sequences using an LRT, comparing the NSsites models M7 (beta) and M8 (beta and ω). The results showed that the M7 model could not be rejected (P = 0.93), and no positively selected sites were reported across the sequences. Taken together, our dN/dS analysis indicate that the seven ItypORs are under strong purifying selection.
ItypOR23 and ItypOR29 Respond to Volatiles Produced by Fungal Symbionts
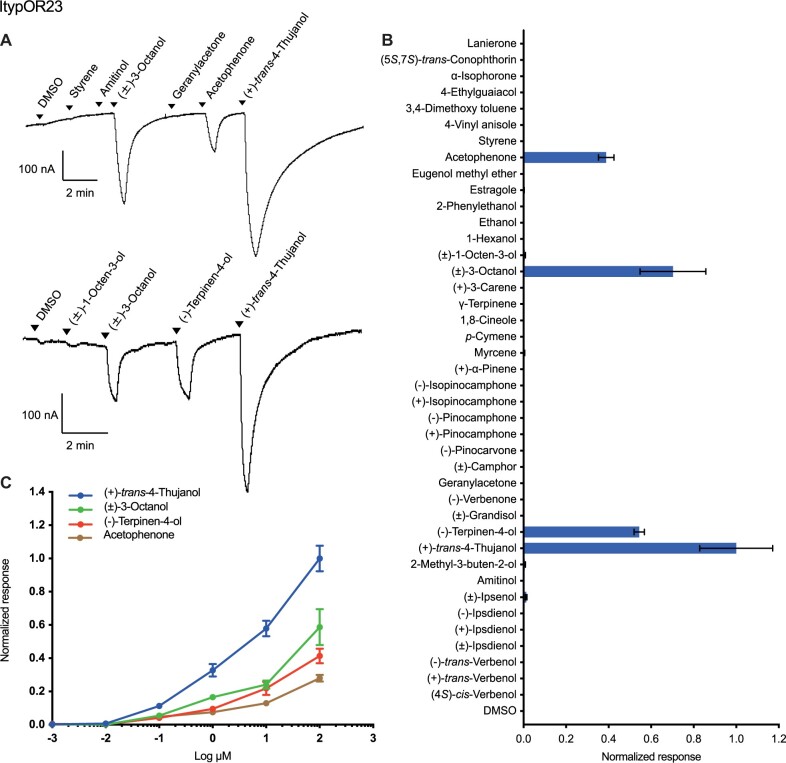
Screening experiments with Xenopus oocytes coexpressing ItypOrco and ItypOR23 showed a primary response to (+)-trans-4-thujanol, and secondary responses to (±)-3-octanol, (–)-terpinen-4-ol, as well as acetophenone (fig. 2A and B). Dose–response experiments showed that ItypOR23 is more sensitive to (+)-trans-4-thujanol than to the other three compounds (fig. 2C), with this compound eliciting larger responses at each tested concentration. Two recent studies (Schiebe et al. 2019; Kandasamy et al. 2021) using partly overlapping odor panels characterized an OSN class specific for (+)-trans-4-thujanol (named OSN class tMTol), with responses very similar to those of ItypOR23 (table 1). Although a weak secondary response to 1-octen-3-ol was observed in the OSN (Kandasamy et al. 2021) but not in the OR, and a weak secondary response to acetophenone in the OR but not in the OSN, the high similarity in the responses to the three best ligands suggests that ItypOR23 is likely to be the matching OR for this OSN class.
Fig. 2.
Responses of ItypOR23 in oocytes. (A) Representative current traces of oocytes upon successive exposures to 100 μM stimuli. Each compound was applied at the time indicated by the arrowheads for 20 s. Upper and lower traces include different sets of test stimuli. (B) Response profile of ItypOR23 to the full odor panel. Values were normalized based on the average response to the primary ligand of ItypOR23 (n ≥ 4). (C) Dose-dependent responses of oocytes expressing ItypOR23 to active ligands. Values were normalized based on the average response of ItypOR23 to the most active compound at 100 μM (n ≥ 4 for each ligand). Error bars indicate the SE.
Table 1.
Comparing in vitro responses of ORs with OSN responses.
| Stimulusa | Matching ORs/OSNs |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 |
2 |
3 |
4c |
|||||
| OR23 | OSN tMTol | OR25b | OSN Δ3 | OR27 | OSN pC | OR29 | OSN Pcn | |
| (+)-trans-4-Thujanol | 1 | 1 | 5 | 2 | 2 | 2 | — | — |
| (–)-Terpinen-4-ol | 2 | 2 | — | — | — | — | — | — |
| (±)-3-Octanol | 2 | 2 | 5 | — | — | — | — | — |
| (±)-1-Octen-3-ol | — | 3 | — | — | — | — | — | — |
| Acetophenone | 3 | — | 5 | 4 | — | — | — | — |
| (+)-3-Carene | — | — | 1 | 1 | — | 2 | — | — |
| (+)-trans-Verbenol | — | — | 2 | — | — | — | — | — |
| Styrene | — | — | 6 | 2 | — | — | — | — |
| 1-Hexanol | — | — | 3 | 3 | — | — | — | — |
| p-Cymene | — | — | — | — | 1 | 1 | — | — |
| γ-Terpinene | — | — | — | — | 1 | 1 | — | — |
| (+)-α-Pinene | — | — | 5 | 4 | — | 2 | — | — |
| Lanierone | — | — | 4 | — | — | — | — | — |
| α-Isophorone | — | — | 4 | — | — | — | — | — |
| (+)-Isopinocamphone | — | 6 | 3 | — | — | 1 | 1 | |
| (–)-Isopinocamphone | — | — | — | — | — | — | 3 | 3 |
| (+)-Pinocamphone | — | — | — | — | — | — | 2 | 2 |
| (–)-Pinocamphone | — | — | — | — | — | — | 3 | 5 |
| (–)-Pinocarvone | — | — | — | — | — | — | 3 | 3 |
| (±)-Camphor | — | — | 6 | 3 | — | — | 4 | 4 |
Note.—Rank order (1 = best compound, 2 = second best, etc.) based on response magnitude (OSN data from Tømmerås [1985]; Andersson et al. [2009]; Schiebe et al. [2019]; Kandasamy et al. [2021]). Normalized responses for ItypORs in oocytes. Repeated number in the same column indicates similar response magnitude.
Only stimuli that were tested on both the OR and the putatively corresponding OSN class are shown.
Additional compounds eliciting minute screening responses in ItypOR25 are not listed for clarity.
Note, apart from (+)-isopinocamphone which stands out as the best ligand, the secondary responses to the structurally related secondary ligands are all very similar; hence, the few discrepancies in compound rank order between the OR and OSNs are minor in terms of sensitivity.
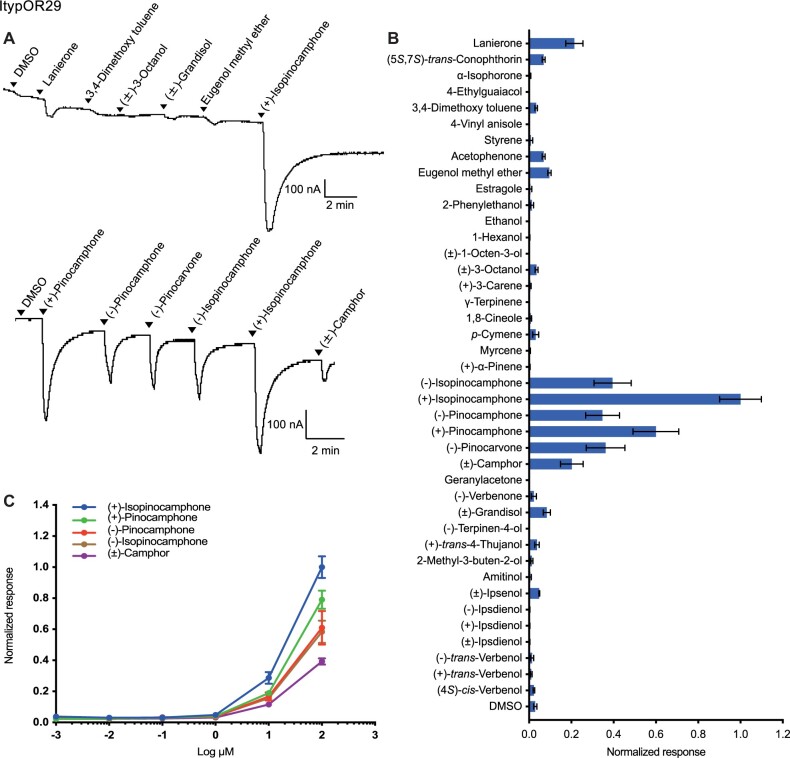
In the screening experiment, oocytes coexpressing ItypOrco and ItypOR29 responded most strongly to (+)-isopinocamphone, followed by rather strong secondary responses to the structurally similar ketones (+)-pinocamphone, (–)-pinocarvone, (–)-pinocamphone, (–)-isopinocamphone, and (±)-camphor (fig. 3A and B). As a result of the thermodynamic equilibrium, both the synthesized pinocamphone enantiomers contained 16–19% impurities of the corresponding isopinocamphone enantiomers (supplementary table S1, Supplementary Material online), and these may have influenced the responses to the pinocamphones. Several additional compounds elicited weaker responses in the screening experiment (fig. 3B). The dose–response assays confirmed that (+)-isopinocamphone is the primary ligand for this OR, although the responses elicited by the secondary ligands were also comparatively high and similar among the tested compounds (fig. 3C). The responses of ItypOR29 are highly similar to the responses of the putatively corresponding OSN class (named OSN class Pcn; Schiebe et al. 2019; Kandasamy et al. 2021), with only minor discrepancies in the rank order among the secondary ligands, which are all similarly active (table 1).
Fig. 3.
Responses of ItypOR29 in oocytes. (A) Representative current traces of oocytes upon successive exposures to 100 μM stimuli. Each compound was applied at the time indicated by the arrowheads for 20 s. Upper and lower traces include different sets of test stimuli. (B) Response profile of ItypOR29 to the full odor panel. Values were normalized based on the average response to the primary ligand of ItypOR29 (n ≥ 4). (C) Dose-dependent responses of oocytes expressing ItypOR29 to active ligands. Values were normalized based on the average response of ItypOR29 to the most active compound at 100 μM (n ≥ 4 for each ligand). Error bars indicate the SE.
ItypOR25 and ItypOR27 Respond to Host Plant Volatiles
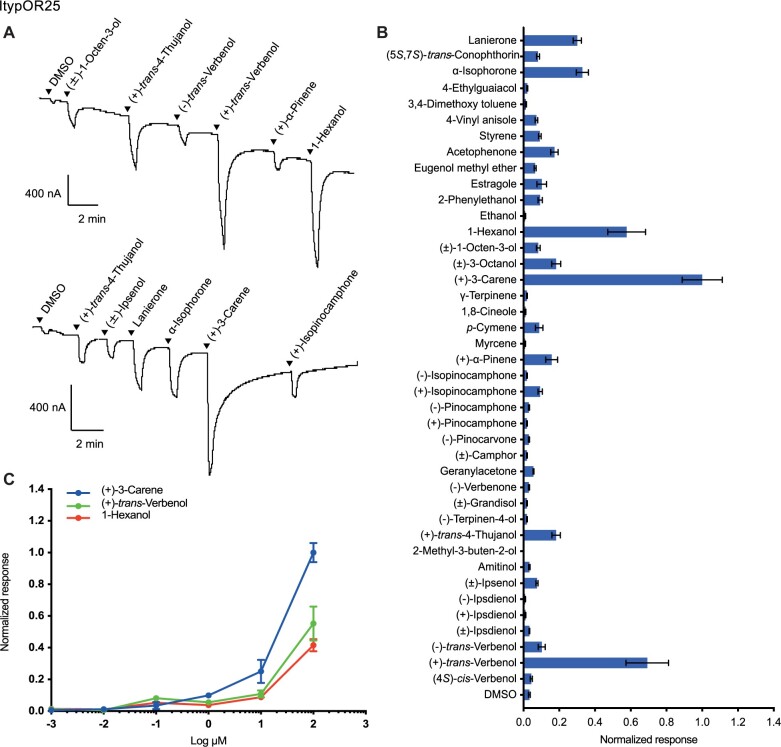
Xenopus oocytes coexpressing ItypOrco and ItypOR25 responded to several compounds in the screening experiment. The strongest response was elicited by the conifer volatile (+)-3-carene followed by the beetle-produced compound (+)-trans-verbenol and the nonhost plant volatile 1-hexanol. Weaker, yet clear, responses were elicited primarily by lanierone and α-isophorone, whereas responses to additional compounds, such as acetophenone, (±)-3-octanol and (+)-trans-4-thujanol, were minor (fig. 4A and B). The dose–response experiments for ItypOR25 indicated a higher sensitivity to (+)-3-carene as compared with the secondary ligands (fig. 4C). Previous SSR experiments characterized an OSN class primarily excited by 3-carene (Andersson et al. 2009; Kandasamy et al. 2021). In addition, several secondary compounds are also shared between this OSN class (named OSN class Δ3; Andersson et al. 2009) and ItypOR25, although the rank orders between them are partially different (table 1).
Fig. 4.
Responses of ItypOR25 in oocytes. (A) Representative current traces of oocytes upon successive exposures to 100 μM stimuli. Each compound was applied at the time indicated by the arrowheads for 20 s. Upper and lower traces include different sets of test stimuli. (B) Response profile of ItypOR25 to the full odor panel. Values were normalized based on the average response to the primary ligand of ItypOR25 (n ≥ 4). (C) Dose-dependent responses of oocytes expressing ItypOR25 to active ligands. Values were normalized based on the average response of ItypOR25 to the most active compound at 100 μM (n ≥ 4 for each ligand). Error bars indicate the SE.
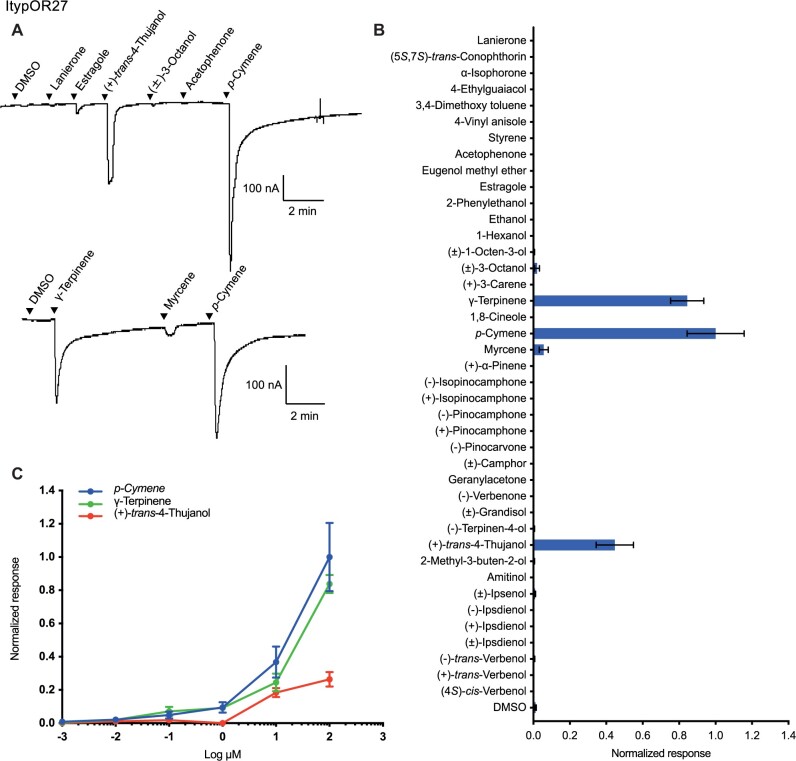
Oocytes coexpressing ItypOrco and ItypOR27 responded strongly and similarly to the two structurally similar host plant volatiles p-cymene and γ-terpinene in the screening assays (fig. 5A and B). A weaker response was elicited by (+)-trans-4-thujanol. Dose–response experiments showed that ItypOR27 displayed a slightly higher sensitivity to p-cymene than to γ-terpinene, and a lower sensitivity to (+)-trans-4-thujanol (fig. 5C). Similar to this OR, the OSN class originally reported to be highly specific for p-cymene (named OSN class pC; Andersson et al. 2009), was recently shown to respond just slightly less to γ-terpinene (Schiebe et al. 2019), and weaker responses to (+)-trans-4-thujanol and additional compounds have also been reported (Kandasamy et al. 2021). Hence, our data for ItypOR27 correspond very well with OSN data reported by Schiebe et al. (2019) (table 1), whereas several of the weakly activating compounds reported by Kandasamy et al. (2021), were inactive in our oocyte experiments.
Fig. 5.
Responses of ItypOR27 in oocytes. (A) Representative current traces of oocytes upon successive exposures to 100 μM stimuli. Each compound was applied at the time indicated by the arrowheads for 20 s. Upper and lower traces include different sets of test stimuli. (B) Response profile of ItypOR27 to the full odor panel. Values were normalized based on the average response to the primary ligand of ItypOR27 (n ≥ 4). (C) Dose-dependent responses of oocytes expressing ItypOR27 to active ligands. Values were normalized based on the average response of ItypOR27 to the most active compound at 100 μM (n ≥ 3 for each ligand). Error bars indicate the SE.
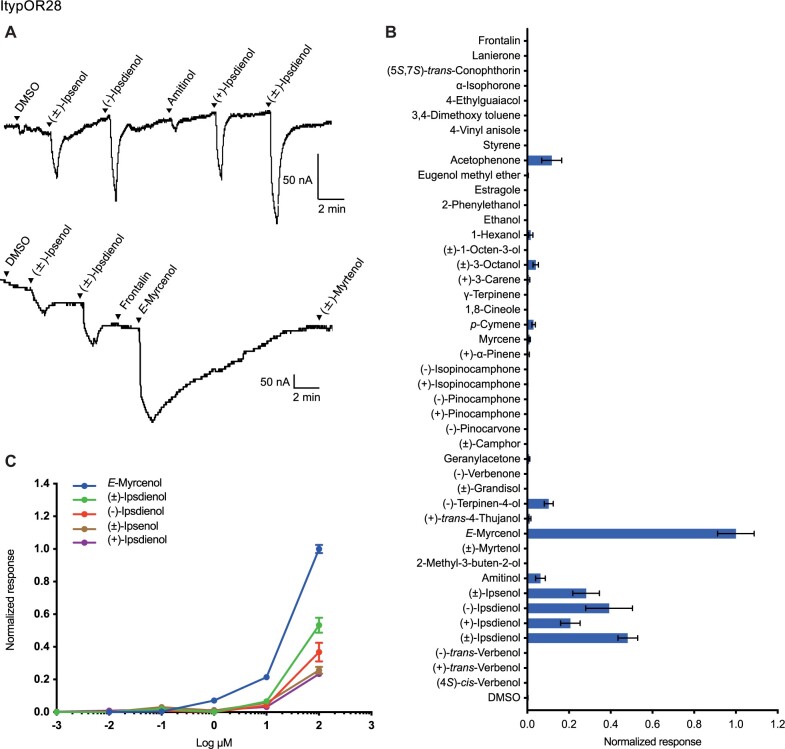
ItypOR28 Responds to a Pheromone Component of the Sympatric I. duplicatus
Oocytes coexpressing ItypOrco and ItypOR28 were activated by bark beetle pheromones, showing the strongest response to E-myrcenol in the screening experiment. E-myrcenol is used as an aggregation pheromone component by the competitor I. duplicatus (Byers et al. 1990). Secondary responses were elicited by the structurally related compounds ipsdienol and ipsenol (fig. 6A and B). Dose–response experiments that included E-myrcenol, racemic ipsenol, racemic ipsdienol, and its two pure enantiomers showed that the response threshold of ItypOR28 is lower for E-myrcenol compared with that for both ipsdienol and ipsenol (fig. 6C). An OSN responding to E-myrcenol in I. typographus has to our knowledge not been reported.
Fig. 6.
Responses of ItypOR28 in oocytes. (A) Representative current traces of oocytes upon successive exposures to 100 μM stimuli. Each compound was applied at the time indicated by the arrowheads for 20 s. Upper and lower traces include different sets of test stimuli. (B) Response profile of ItypOR28 to the full odor panel. Values were normalized based on the average response to the primary ligand of ItypOR28 (n ≥ 4). (C) Dose-dependent responses of oocytes expressing ItypOR28 to active ligands. Values were normalized based on the average response of ItypOR28 to the most active compound at 100 μM (n ≥ 4 for each ligand). Error bars indicate the SE.
Discussion
Functional Evolution of the OR-Radiation
A recent phylogenetic analysis (Yuvaraj et al. 2021) that included the 73 antennally expressed ORs of I. typographus along with the ORs from the mountain pine beetle D. ponderosae (Andersson et al. 2019) and the Asian longhorn beetle Anoplophora glabripennis (Cerambycidae) (McKenna et al. 2016) revealed a highly supported clade formed by seven ItypORs. Two of these receptors (ItypOR46 and ItypOR49) were shown to selectively respond to the pheromone compounds (S)-(–)-ipsenol (ItypOR46) and (R)-(–)-ipsdienol (ItypOR49) (Yuvaraj et al. 2021). Here, we obtained functional data for the five remaining ItypORs from this clade. Collectively, our results show that this clade contains two ORs specifically tuned to monoterpene hydrocarbons produced by the host tree (ItypOR25 and ItypOR27), three ORs tuned to oxygenated monoterpenoids produced by con- or heterospecific bark beetles (i.e., pheromone compounds; ItypOR28, ItypOR46, and ItypOR49), and two ORs responding to volatiles produced by the symbiotic fungi of I. typographus (ItypOR23 and ItypOR29). Similar to previous suggestions (Mitchell et al. 2012; Mitchell and Andersson 2020; Yuvaraj et al. 2021), these findings strengthen the idea that pheromone receptors (PRs) from different beetle species do not cluster in specific clades like PRs do in Lepidoptera where the majority of the characterized PRs are found in the so-called classical PR clade (Zhang and Löfstedt 2015). Instead, beetle PRs are scattered in the OR phylogeny, and OR clades include receptors detecting compounds from various ecological sources. Specifically, among the nine major monophyletic groups of coleopteran ORs (Mitchell et al. 2020), the three I. typographus PRs belong to Group 7, which is expanded in curculionids. The PR RferOR1 from the red palm weevil Rhynchophorus ferrugineus (also Curculionidae) is also part of Group 7 but positioned in a distantly related OR subfamily (Antony et al. 2021). In contrast, the three PRs in the cerambycid Megacyllene caryae are found in OR Group 1 (McarOR20) and 2B (McarOR3 and 5) (Mitchell et al. 2012), hence phylogenetically very distantly. Similar to the beetle PRs, ORs in ants that detect cuticular hydrocarbons (contact pheromones) are present in numerous phylogenetically distinct subfamilies, both within and outside the so-called nine-exon OR subfamily specific to Hymenoptera (McKenzie et al. 2016; Pask et al. 2017; Slone et al. 2017).
The grouping of bark beetle PRs with ORs detecting compounds from other ecological sources may not be surprising because most bark beetle pheromones are oxygenated monoterpenoids, sometimes produced through metabolism of host monoterpenes, and hence structurally similar to common conifer host tree compounds (and to several fungal-derived compounds). This contrasts the situation in moths where most pheromone compounds are structurally different from plant volatiles. In addition, host- and mate localization in bark beetles is an integrated process involving pheromones, host and nonhost odors, with aggregation pheromones mediating sex-attraction, aggregation behavior, and successful localization and infestation of host trees providing food and oviposition sites.
Although our functional data suggest a clade in which all ItypORs detect monoterpenes or monoterpenoids and are under strong purifying selection, our findings also bring up questions of which odor specificities may evolve in OR-radiations. Whereas the pheromone compounds ipsenol, ipsdienol, and E-myrcenol are similar acyclic monoterpenols with a myrcene backbone, the other ligands for the ORs in this clade include monoterpene hydrocarbons and ketones as well as cyclic structures (fig. 1D). The splits between the seven ORs appear to be rather old, as indicated by the branch lengths (fig. 1A) and low amino acid identities (fig. 1C), and this appears to have provided sufficient time for significant specificity shifts (neofunctionalization) to evolve. Purifying selection against deleterious mutations then appears to have dominated to retain the OR responses to these ecologically important compounds. Under such a scenario, it is difficult to infer which specific amino acid changes have resulted in the observed specificity shifts. Clades housing more recently diverged ORs should be investigated to inform this question; however, such OR clades have so far not been identified in I. typographus.
Previous work (Tømmerås et al. 1984; Andersson et al. 2009) identified OSNs primarily responding to, for example, (S)-(+)-ipsdienol and amitinol, which are both structurally related to the primary ligands (S)-(–)-ipsenol, (R)-(–)-ipsdienol, and E-myrcenol of ORs characterized here. Unexpectedly, the ORs detecting these two similar compounds are not found within this receptor lineage, and hence must have evolved their specificities independently. The same holds true for the several OSN classes specifically tuned to monoterpene hydrocarbons or oxygenated monoterpenoids (Andersson et al. 2009; Schiebe et al. 2019), of which only four putatively corresponding ORs are localized in this clade. Across several species of Lepidoptera, the structures of key ligands for characterized ORs follow the OR phylogeny at a broader scale, with ligands detected by relatively closely related ORs sharing similar molecular features (de Fouchier et al. 2017; Guo et al. 2021). For instance, the ORs within the classical PR clade are typically tuned to long-chain fatty acid derivatives, and other OR clades also house receptors that primarily respond to a specific class of chemicals, such as aromatics or terpenes (de Fouchier er al. 2017). Similar to our findings there is, however, also variation within these clades with respect to, for example, functional groups, chain length, branching, and unsaturation among ligands detected by the related ORs. Likewise, the Drosophila melanogaster ORs DmelOR22a, 43b, 59b, 85a, and 98a, which belong to the same OR clade respond primarily to esters (Hallem and Carlson 2006; Mansourian and Stensmyr 2015). However, a few additional ORs in this expansion do not have an ester as the key ligand, and the branch lengths in the OR clade indicate that the splits are old (Robertson et al. 2003). In the Anopheles gambiae mosquito, the related ORs AgamOR13, 15, 16, and 18 are activated by aromatics, and with overlapping responses to, for example, acetophenone and benzaldehyde (Carey et al. 2010; Wang et al. 2010). This suggests that subfunctionalization (rather than neofunctionalization) has operated on these particular ORs.
Conversely, studies have also shown that phylogenetically widely separated ORs may be tuned to structurally similar compounds. For example, the recently identified “novel” PR clade in Lepidoptera harbors receptors responsive to structural isomers of the ligands for the receptors in classical PR clade (Zhang and Löfstedt 2015), although the two clades are distant (Bastin-Héline et al. 2019). It was suggested that Lepidoptera PRs have evolved independently multiple times, although most of them reside in the classical PR clade. Also in ants, several of the ORs that detect cuticular hydrocarbons that only differ slightly in chain length are widely separated in the OR phylogeny (Pask et al. 2017; Slone et al. 2017).
Apart from the model dipterans D. melanogaster and A. gambiae, our study represents a rare example where all OR paralogues within a clade comprising as many as seven receptors from one species have been functionally characterized. Hence, large-scale functional characterization of the OR repertoires of I. typographus and additional species is required to further our understanding of the functional evolution of the insect OR gene family.
ItypOR23 and ItypOR29 Detect Volatiles Produced by Fungal Symbionts
Like other bark beetles, I. typographus is associated with a suite of symbiotic ophiostomatoid fungi, which beetles inoculate into the inner bark of attacked trees. These fungi may serve as food for maturing beetles (Kandasamy et al. 2019) and they also metabolize spruce defense compounds (Kandasamy et al. 2021). In laboratory bioassays, beetles actively seek out fungal-colonized media using olfactory cues and are able to distinguish different fungal species by their different odor profiles using OSNs dedicated for fungal volatiles (Kandasamy et al. 2019). Here, we characterized two receptors, ItypOR23 and ItypOR29, responding to the fungal-derived volatiles (+)-trans-4-thujanol and (+)-isopinocamphone, respectively. Both of these ORs showed high response specificities to only a few structurally related compounds. The OR response profiles match very well with those from the characterized OSNs, where the tMTol class is highly specific for (+)-trans-4-thujanol and the Pcn class selective for the highly similar monoterpene ketones that activate ItypOR29 (table 1; Schiebe et al. 2019; Kandasamy et al. 2021).
It was recently shown that the fungi of I. typographus transform the host compounds (–)-β-pinene, (–)-α-pinene and (–)-bornyl acetate into oxygenated monoterpenoids, including the primary and secondary ligands of both ItypOR23 and ItypOR29 (Kandasamy et al. 2021). In lab bioassays, (+)-trans-4-thujanol and camphor were attractive to adult beetles at a relatively low dose (100 µg), however, (+)-trans-4-thujanol was repellent at higher doses (200 µg to 1 mg) (Blažytė-Čereškienė et al. 2016; Kandasamy et al. 2021). This suggests concentration-dependent effects of these compounds and that changes in their emission over time may be used by new incoming bark beetles to evaluate the colonization status of the host tree. In addition to production by fungi, these oxygenated monoterpenoids are also released from spruce trees, with amounts decreasing with tree age and increasing with tree stress (Blažytė-Čereškienė et al. 2016; Schiebe et al. 2019). Hence, these compounds of multiple ecological origins may be also used by I. typographus to assess the physiological status of host trees.
ItypOR25 and ItypOR27 Are Tuned to Host Plant Volatiles
ItypOR25 and OR27 were most sensitive to two host monoterpenes, (+)-3-carene and p-cymene, respectively, and especially ItypOR27 demonstrated a high specificity. These two ORs group together within the analyzed OR clade and present the most recent split among the seven ORs, which is in line with a tuning shift from detecting oxygenated monoterpenoids to monoterpene hydrocarbons. Despite this shared characteristic, the response profiles of the two ORs are widely different suggesting that neofunctionalization has been favored also after their presumed duplication (Rastogi and Liberles 2005; Andersson et al. 2015). Whereas the response profile of ItypOR27 matches very well with that of the pC OSN class, ItypOR25 presented a worse match with a broader response profile and partly different rank orders among the secondary compounds compared with the putatively corresponding Δ3 OSN class (table 1; Andersson et al. 2009; Schiebe et al. 2019). It remains unknown whether this discrepancy is due to the artificial oocyte environment or whether ItypOR25 is not the receptor of the Δ3 OSN class. However, differences in OR responses between different in vitro systems and between these systems and the OSNs have repeatedly been found in previous studies, including for ItypOR46 and ItypOR49 and corresponding OSN classes (Yuvaraj et al. 2017; Hou et al. 2020; Yuvaraj et al. 2021). Because of this system dependency and because no other OSN in I. typographus has shown a primary response to 3-carene (Andersson et al. 2009; Kandasamy et al. 2019; Schiebe et al. 2019), it appears likely that ItypOR25 is indeed the receptor that corresponds to the Δ3 OSN class.
The roles of host monoterpenes in host selection by I. typographus is poorly understood and likely context dependent. Although attraction to host monoterpenes alone has not been demonstrated, the major host compounds α‐pinene and β-pinene can enhance the response of the bark beetle to the aggregation pheromone (Rudinsky et al. 1972; Hulcr et al. 2006; Erbilgin et al. 2007). In contrast, less abundant host compounds may reduce the pheromone attraction. For example, 1,8-cineole is more abundant in resistant trees and upregulated during an attack, and this compound has a strong antagonistic effect on pheromone attraction (Andersson et al. 2010; Schiebe et al. 2012; Binyameen et al. 2014). Similarly, p-cymene is increased in heavily attacked trees, and it reduced pheromone trap catch by 50% (Andersson et al. 2010). Hence, increased amounts of inducible toxic host compounds may be used by bark beetles as cues to evaluate host defense potential and/or breeding density, and may therefore represent general “host unsuitability” signals (Erbilgin and Raffa 2000; Seybold et al. 2006; Andersson et al. 2010) with context- and species-dependent effects (Saint-Germain et al. 2007; Raffa et al. 2016). To our knowledge, behavioral effects of 3-carene have not been studied in I. typographus. However, the amount of 3-carene in Norway spruce increases after inoculation with the I. typographus-associated fungus Endoconidiophora polonica, and this compound has been linked to conifer resistance and susceptibility to insects and fungi (Zhao et al. 2010, and references therein). In addition, 3-carene is also abundant in pine trees, including sympatric Scots pine (Pinus sylvestris) and it may hence be used as a host suitability cue, similar to p-cymene and 1,8-cineole.
ItypOR28, ItypOR46, and ItypOR49 Detect Bark Beetle Pheromones
We characterized one bark beetle PR (ItypOR28) primarily responding to E-myrcenol. Hence, together with the previously characterized ItypOR46 ((S)-(–)-ipsenol) and ItypOR49 ((R)-(–)-ipsdienol) (Yuvaraj et al. 2021), the targeted clade houses three PRs. The ipsenol and ipsdienol enantiomers are produced by male I. typographus during the later attack phases (2–6 days after boring has been initiated), and at least ipsenol inhibits the attraction to the aggregation pheromone, possibly to avoid intraspecific competition (Francke et al. 1980; Birgersson et al. 1984; Schlyter et al. 1989). E-myrcenol together with both enantiomers of ipsdienol comprise the aggregation pheromone of I. duplicatus, the largest competitor of I. typographus (Byers et al. 1990), and E-myrcenol inhibits the attraction of I. typographus to its aggregation pheromone (Schlyter et al. 1992). Consistent with a general high specificity of insect PRs (Andersson et al. 2015), ItypOR28 only responded secondarily to the structurally related ipsdienol and ipsenol, and narrow tunings were also reported for ItypOR46 and ItypOR49 (Yuvaraj et al. 2021). In contrast to the four other ItypORs characterized here, no OSN class has been reported to be specific for E-myrcenol despite large screening efforts that included this compound (Andersson et al. 2009). Our results from ItypOR28 suggest the existence of such an OSN class, and it is possible that these OSNs have been missed because some OSN classes were recently shown to have a highly restricted spatial distribution on the I. typographus antenna (Kandasamy et al. 2019).
Concluding Remarks
We determined the functions of five ORs from an Ips-specific OR clade using the Xenopus oocyte system. Together with the two PRs previously characterized in HEK cells and oocytes (Yuvaraj et al. 2021), this clade contains seven ORs which respond to monoterpenes or monoterpenoids, indicating the biological importance of such compounds to I. typographus. Three ORs respond to bark beetle pheromones, two ORs to host plant volatiles, and two ORs to compounds produced in relatively large amounts by fungal symbionts. The structural similarities especially among the three pheromonal ligands detected by ItypOR28, ItypOR46, and ItypOR49 shows that several ORs in this radiation have evolved specificities for chemically similar compounds. In addition, the variation in the structures of the key ligands for the seven receptors and their various ecological origins suggest that neofunctionalization has been promoted in the evolution of this OR clade after which purifying selection has dominated to retain the new OR responses. Finally, our results indicate that the PRs of beetles do not form specific PR clades as seen in Lepidoptera (Yuvaraj et al. 2018). Instead, beetle PRs are dispersed across the OR phylogeny, and OR-lineage radiations within species contain receptors that detect compounds of different ecological origins.
Materials and Methods
Chemicals
Compounds tested in this study were from commercial sources, synthesized by the Unelius laboratory, or obtained from colleagues as gifts (supplementary table S1, Supplementary Material online). The test compounds included beetle-produced pheromone compounds, and volatiles produced by associated fungi, conifer host and angiosperm nonhost trees, including the key ligands for all known OSN classes of I. typographus as well as several secondary ligands for these neurons (Tømmerås 1985; Andersson et al. 2009; Kandasamy et al. 2019; Schiebe et al. 2019; Kandasamy et al. 2021). Stock solutions for Xenopus oocyte recordings were prepared by diluting each compound to 100 mM in dimethyl sulfoxide (DMSO), which were stored at −20 °C. Before each experiment, the stock solutions were diluted to desired concentration in Ringeŕs buffer (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, 0.8 mM CaCl2, 5 mM HEPES, pH 7.6) with the final stimuli containing 0.1% DMSO. Ringeŕs buffer containing 0.1% DMSO was used as negative control. Compounds were initially screened for receptor activity at a concentration of 100 μM, and the active compounds were subsequently tested in dose–response experiments using six concentrations at 10-fold increments ranging from 1 nM to 100 µM. An odor panel of 32 compounds was initially tested on all five ORs (ItypOR23, 25, 27, 28, and 29) (supplementary table S1, Supplementary Material online). After obtaining the first screening responses to this odor panel, additional OR-specific compounds (putative secondary ligands) were selected for testing based on previous data from putatively matching OSN classes, which facilitated our comparisons of OR responses with those of previously characterized OSNs (supplementary table S2, Supplementary Material online). For consistency, these nine additional compounds were also tested on the other nontarget ORs over two replications (showing insignificant activity). For ItypOR28, the initial screening experiment indicated only minor and rather unspecific responses to some test compounds. Hence, we hypothesized that this OR may be tuned to a structurally related compound, and therefore also tested frontalin, E-myrcenol and myrtenol specifically on this OR (supplementary table S3, Supplementary Material online). Chemical structures presented in figure 1D were drawn using ChemDraw Professional 17.0.
Vector Construction and cRNA Synthesis
Full-length sequences of ItypORs and ItypOrco were amplified by PCR using gene-specific primers containing flanking 5′ Kozak sequence (“GCCACC”) and 5′ and 3′ recognition sites for restriction enzymes (supplementary table S4, Supplementary Material online). The expression vectors pcDNA5/TO containing the five ItypORs (ItypOR23, 25, 27, 28, and 29; amino acid sequences are available in the supplementary material, Supplementary Material online) and pcDNA4/TO containing ItypOrco that were used in previous HEK293 cell assays (Yuvaraj et al. 2021) were used as templates for subcloning. The PCR products were analyzed on 1% TAE agarose gels, and then purified using the GeneJET Gel Extraction Kit (Thermo Fisher Scientific). The purified fragments were ligated into the oocyte expression vector pCS2+ using T4 ligase (Thermo Fisher Scientific) overnight at 4 °C and then transformed into TOP10 competent cells (Thermo Fisher Scientific). Positive colonies that were confirmed by colony PCR were grown in LB broth overnight with ampicillin, and the plasmids were then extracted using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific). Plasmids were Sanger sequenced on a capillary 3,130 × l Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) using Sp6 and EBV-rev primers at the sequencing facility at the Department of Biology, Lund University. Large quantities of purified plasmids containing ItypORs and ItypOrco with correct sequences were obtained using the PureLinkTM HiPure Plasmid Filter Midiprep Kit (Thermo Fisher Scientific). cRNAs of ItypORs and ItypOrco were synthesized from NotI (Promega) linearized recombinant pCS2+ plasmids using the mMESSAGE mMACHINE SP6 transcription kit (Thermo Fisher Scientific). Sequences of the five ItypORs have been deposited in GenBank under the accession numbers MW556722-MW556726.
Microinjection and Two-Electrode Voltage Clamp Recordings
Each of the five ItypORs was coexpressed with ItypOrco in Xenopus laevis oocytes, and two-electrode voltage clamp recordings were performed following previously described protocols (Zhang et al. 2010; Zhang and Löfstedt 2013). In brief, oocytes were surgically collected from X. laevis females (frogs were purchased from University of Portsmouth, UK) and treated with 1.5 mg/ml collagenase (Sigma-Aldrich Co., St Louis, MO, USA) in oocyte Ringer 2 buffer (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES, pH 7.5) at room temperature for 15–18 min. Mature healthy oocytes (stages V–VII) were coinjected with 40 ng each of candidate OR and Orco cRNAs, then incubated for 3–7 days at 18 °C in Ringer’s buffer containing sodium pyruvate (550 mg/l) and gentamicin (100 mg/l). Odorant-induced whole-cell inward currents were recorded from injected oocytes in good condition using a two-electrode voltage clamp with a TEC-03BF amplifier at the holding potential of −80 mV. A computer-controlled perfusion system was employed to apply the tested compounds and Ringer’s buffer to the oocyte chamber. Oocytes were exposed to compounds at a flow rate of 2 ml/min for 20 s with extensive washing with Ringer’s buffer at a flow rate of 4 ml/min between stimulations to recover the current to baseline. Data were collected and analyzed using the Cellworks software (NPI Electronic GmbH, Tamm, Germany), and the responses were normalized by calculating the relative response to each active compound in relation to the average response to the most active compound.
Evolutionary Analysis of ORs
Amino acid sequences of select Group 7 ORs (Mitchell et al. 2020) from I. typographus and D. ponderosae were aligned using MAFFT (v7.450; Katoh et al. 2002), and an unrooted maximum-likelihood tree was constructed using FastTree 2.1.11 (Price et al. 2010) implemented in Geneious Prime (2020.0.5) software (Biomatters Ltd., Auckland, New Zealand). Local node support values were calculated using the Shimodaira–Hasegawa (SH) test implemented in FastTree. The nonsynonymous (dN) to synonymous (dS) substitution rate (ω) was estimated by the maximum-likelihood method (Anisimova et al. 2001) using the Codeml program in the PAML 4.6 package (Yang 1997).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to Yoko Nakamura, Suresh Ganji, Erika Wallin, Blanka Kalinová, Fredrik Schlyter, Anna Jirošová, and Aleš Machara for providing test compounds. We thank the Chinese Scholarship Council (CSC) for finical support of Xiaoqing Hou’s PhD study. This work was also supported by grants from the Swedish Research Councils FORMAS (Grant Nos. 217-2014-689, 2018-01444 to M.N.A.) and VR (Grant No. 2017-03804 to C.L.), the Crafoord foundation (to M.N.A.), the Carl Trygger Foundation (Grant No. CTS 17:25 to M.N.A.), and the Royal Physiographic Society in Lund (to R.E.R. and M.N.A.). The study was carried out as part of the Max Planck Centre for Next Generation Chemical Ecology (nGICE). AQ7
Data Availability
Data from the oocyte recordings are presented in the main figures of the article. Sequences of the characterized ItypORs have been deposited in GenBank (accession numbers: MW556722–MW556726).
References
- Adipietro KA, Mainland JD, Matsunami H.. 2012. Functional evolution of mammalian odorant receptors. PLoS Genet. 8(7):e1002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN. 2012. Mechanisms of odor coding in coniferous bark beetles: from neuron to behavior and application. Psyche J Entomol. 2012:149572. [Google Scholar]
- Andersson MN, Grosse-Wilde E, Keeling CI, Bengtsson JM, Yuen MM, Li M, Hillbur Y, Bohlmann J, Hansson BS, Schlyter F.. 2013. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics. 14:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN, Keeling CI, Mitchell RF.. 2019. Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genomics. 20(1):690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MN, Larsson MC, Blaženec M, Jakuš R, Zhang QH, Schlyter F.. 2010. Peripheral modulation of pheromone response by inhibitory host compound in a beetle. J Exp Biol. 213(Pt 19):3332–3339. [DOI] [PubMed] [Google Scholar]
- Andersson MN, Larsson MC, Schlyter F.. 2009. Specificity and redundancy in the olfactory system of the bark beetle Ips typographus: single-cell responses to ecologically relevant odors. J Insect Physiol. 55(6):556–567. [DOI] [PubMed] [Google Scholar]
- Andersson MN, Löfstedt C, Newcomb RD.. 2015. Insect olfaction and the evolution of receptor tuning. Front Ecol Evol. 3:53. [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z.. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 18(8):1585–1592. [DOI] [PubMed] [Google Scholar]
- Antony B, Johny J, Montagné N, Jacquin-Joly E, Capoduro R, Cali K, Persaud K, Al-Saleh MA, Pain A.. 2021. Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus). Mol Ecol. 30(9):2025–2039. [DOI] [PubMed] [Google Scholar]
- Bastin-Héline L, De Fouchier A, Cao S, Koutroumpa F, Caballero-Vidal G, Robakiewicz S, Monsempes C, François MC, Ribeyre T, Maria A.. 2019. A novel lineage of candidate pheromone receptors for sex communication in moths. Biochem Pharmacol. 24(17):e49826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann PHW, Müller J, Grégoire JC, Gruppe A, Hagge J, Hammerbacher A, Hofstetter R, Kandasamy D, Kolarik M, Kostovcik M, et al. 2019. Bark beetle population dynamics in the Anthropocene: challenges and solutions. Trends Ecol Evol. 34(10):914–924. [DOI] [PubMed] [Google Scholar]
- Binyameen M, Jankuvová J, Blaženec M, Jakuš R, Song L, Schlyter F, Andersson MN.. 2014. Co-localization of insect olfactory sensory cells improves the discrimination of closely separated odor sources. Funct Ecol. 28(5):1216–1223. [Google Scholar]
- Birgersson G, Schlyter F, Löfqvist J, Bergström G.. 1984. Quantitative variation of pheromone components in the spruce bark beetle Ips typographus from different attack phases. J Chem Ecol. 10(7):1029–1055. [DOI] [PubMed] [Google Scholar]
- Blažytė-Čereškienė L, Apšegaitė V, Radžiutė S, Mozūraitis R, Būda V, Pečiulytė D.. 2016. Electrophysiological and behavioural responses of Ips typographus (L.) to trans-4-thujanol—a host tree volatile compound. Ann for Sci. 73(2):247–256. [Google Scholar]
- Byers JA, Schlyter F, Birgersson G, Francke W.. 1990. E-myrcenol in Ips duplicatus: an aggregation pheromone component new for bark beetles. Experientia 46(11–12):1209–1211. [Google Scholar]
- Carey AF, Wang G, Su CY, Zwiebel LJ, Carlson JR.. 2010. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464(7285):66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR.. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22(2):327–338. [DOI] [PubMed] [Google Scholar]
- de Fouchier A, Walker WB, Montagné N, Steiner C, Binyameen M, Schlyter F, Chertemps T, Maria A, François M-C, Monsempes C, et al. 2017. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 8:15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds RL, Eglitis A.. 1989. The role of the Douglas-fir beetle and wood borers in the decomposition of and nutrient release from Douglas-fir logs. Can J for Res. 19(7):853–859. [Google Scholar]
- Erbilgin N, Krokene P, Kvamme T, Christiansen E.. 2007. A host monoterpene influences Ips typographus (Coleoptera: curculionidae Scolytinae) responses to its aggregation pheromone. Agric Forest Ent. 9(2):135–140. [Google Scholar]
- Erbilgin N, Raffa KF.. 2000. Opposing effects of host monoterpenes on responses by two sympatric species of bark beetles to their aggregation pheromones. J Chem Ecol. 26(11):2527–2548. [Google Scholar]
- Fleischer J, Pregitzer P, Breer H, Krieger J.. 2018. Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol Life Sci. 75(3):485–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke W, Sauerwein P, Vité JP, Klimetzek D.. 1980. The pheromone bouquet of Ips amitinus. Naturwissenschaften 67(3):147–148. [Google Scholar]
- Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J.. 2007. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 25(8):2364–2373. [DOI] [PubMed] [Google Scholar]
- Guo M, Du L, Chen Q, Feng Y, Zhang J, Zhang X, Tian K, Cao S, Huang T, Jacquin-Joly E, et al. 2021. Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol Biol Evol. 38(4):1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR.. 2006. Coding of odors by a receptor repertoire. Cell 125(1):143–160. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC.. 2011. Evolution of insect olfaction. Neuron 72(5):698–711. [DOI] [PubMed] [Google Scholar]
- Hopf TA, Morinaga S, Ihara S, Touhara K, Marks DS, Benton R.. 2015. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 6:6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horntvedt R, Christiansen E, Solheim H, Wang S.. 1983. Artificial inoculation with Ips typographus-associated blue stain fungi can kill healthy Norway spruce trees. Medd nor Inst Skogforsk. 38:1–20. [Google Scholar]
- Hou X, Zhang D-D, Yuvaraj JK, Corcoran JA, Andersson MN, Löfstedt C.. 2020. Functional characterization of odorant receptors from the moth Eriocrania semipurpurella: a comparison of results in the Xenopus oocyte and HEK cell systems. Insect Biochem Mol Biol. 117:103289. [DOI] [PubMed] [Google Scholar]
- Hulcr J, Ubik K, Vrkoc J.. 2006. The role of semiochemicals in tritrophic interactions between the spruce bark beetle Ips typographus, its predators and infested spruce. J Appl Entomology. 130(5):275–283. [Google Scholar]
- Kandasamy D, Gershenzon J, Andersson MN, Hammerbacher A.. 2019. Volatile organic compounds influence the interaction of the Eurasian spruce bark beetle (Ips typographus) with its fungal symbionts. Isme J. 13(7):1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy D, Zaman R, Nakamura Y, Zhao T, Hartmann H, Andersson MN, Hammerbacher A, Gershenzon J.. 2021. Bark beetles locate fungal symbionts by detecting volatile fungal metabolites of host tree resin monoterpenes. bioRxiv preprint. doi:10.1101/2021.07.03.450988. [DOI] [PMC free article] [PubMed]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausrud KL, Grégoire JC, Skarpaas O, Erbilgin N, Gilbert M, Økland B, Stenseth NC.. 2011. Trees wanted—dead or alive! Host selection and population dynamics in tree-killing bark beetles. PLoS One. 6(5):e18274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Macallister IE, Kavanaugh MP, Wanner KW.. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci U S A. 109(35):14081–14086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S, Stensmyr MC.. 2015. The chemical ecology of the fly. Curr Opin Neurobiol. 34:95–102. [DOI] [PubMed] [Google Scholar]
- McKenna DD, Scully ED, Pauchet Y, Hoover K, Kirsch R, Geib SM, Mitchell RF, Waterhouse RM, Ahn S-J, Arsala D, et al. 2016. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 17(1):227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SK, Fetter-Pruneda I, Ruta V, Kronauer DJC.. 2016. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc Natl Acad Sci U S A. 113(49):14091–14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RF, Andersson MN.. 2020. Olfactory genomics of the Coleoptera. In: Blomquist GJ, Vogt RG, editors. Insect pheromone biochemistry and molecular biology. 2nd ed. Oxford (UK): Academic Press, p. 547–590. [Google Scholar]
- Mitchell RF, Hughes DT, Luetje CW, Millar JG, Soriano-Agatón F, Hanks LM, Robertson HM.. 2012. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem Mol Biol. 42(7):499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RF, Schneider TM, Schwartz AM, Andersson MN, McKenna DD.. 2020. The diversity and evolution of odorant receptors in beetles (Coleoptera). Insect Mol Biol. 29(1):77–91. [DOI] [PubMed] [Google Scholar]
- Mustaparta H, Tømmerås BÅ, Baeckström P, Bakke JM, Ohloff G.. 1984. Ipsdienol-specific receptor cells in bark beetles: structure–activity relationships of various analogues and of deuterium-labelled ipsdienol. J Comp Physiol. 154(4):591–596. [Google Scholar]
- Nei M, Niimura Y, Nozawa M.. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 9(12):951–963. [DOI] [PubMed] [Google Scholar]
- Pask GM, Slone JD, Millar JG, Das P, Moreira JA, Zhou X, Bello J, Berger SL, Bonasio R, Desplan C, et al. 2017. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat Commun. 8(1):297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2010. FastTree2 - approximately maximum-likely hood trees for large alignments. PLoS One. 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa KF. 2001. Mixed messages across multiple trophic levels: the ecology of bark beetle chemical communication systems. Chemoecology 11:49–65. [Google Scholar]
- Raffa KF, Andersson MN, Schlyter F.. 2016. Host selection by bark beetles: playing the odds in a high-stakes game. In: Tittiger C, Blomquist GJ, editors. Advances in insect physiology. Vol 50. Oxford (UK: ): Academic Press. p. 1–74. [Google Scholar]
- Ramdya P, Benton R.. 2010. Evolving olfactory systems on the fly. Trends Genet. 26(7):307–316. [DOI] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA.. 2005. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR.. 2003. Molecular evolution of the insect chemoreceptor gene subfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100(Suppl 2):14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinsky JA, Furniss MM, Kline LN, Schmitz RF.. 1972. Attraction and repression of Dendroctonus pseudotsugae (Coleoptera: Scolytidae) by three synthetic pheromones in traps in Oregon and Idaho. Can Entomol. 104(6):815–822. [Google Scholar]
- Saint-Germain M, Buddle CM, Drapeau P.. 2007. Primary attraction and random landing in host-selection by wood-feeding insects: a matter of scale? Agric Forest Ent. 9(3):227–235. [Google Scholar]
- Schiebe C, Hammerbacher A, Birgersson G, Witzell J, Brodelius PE, Gershenzon J, Hansson BS, Krokene P, Schlyter F.. 2012. Inducibility of chemical defenses in Norway spruce bark is correlated with unsuccessful mass attacks by the spruce bark beetle. Oecologia 170(1):183–198. [DOI] [PubMed] [Google Scholar]
- Schiebe C, Unelius CR, Ganji S, Binyameen M, Birgersson G, Schlyter F.. 2019. Styrene, (+)-trans-(1R,4S,5S)-4-thujanol and oxygenated monoterpenes related to host stress elicit strong electrophysiological responses in the bark beetle Ips typographus. J Chem Ecol. 45(5–6):474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlyter F, Birgersson G, Byers JA, Bakke A.. 1992. The aggregation pheromone of Ips duplicatus and its role in competitive interactions with I typographus (Coleoptera: Scolytidae). Chemoecology 3(3–4):103–112. [Google Scholar]
- Schlyter F, Birgersson G, Byers JA, Löfqvist J, Bergström G.. 1987. Field response of spruce bark beetle, Ips typographus, to aggregation pheromone candidates. J Chem Ecol. 13(4):701–716. [DOI] [PubMed] [Google Scholar]
- Schlyter F, Birgersson GR, Leufvén A.. 1989. Inhibition of attraction to aggregation pheromone by verbenone and ipsenol. J Chem Ecol. 15(8):2263–2277. [DOI] [PubMed] [Google Scholar]
- Seybold S, Huber D, Lee J, Graves AD, Bohlmann J.. 2006. Pine monoterpenes and pine bark beetles: a marriage of convenience for defense and chemical communication. Phytochem Rev. 5(1):143–178. [Google Scholar]
- Slone JD, Pask GM, Ferguson ST, Millar JG, Berger SL, Reinberg D, Liebig J, Ray A, Zwiebel LJ.. 2017. Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proc Natl Acad Sci U S A. 114(32):8586–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tømmerås BÅ. 1985. Specialization of the olfactory receptor cells in the bark beetle Ips typographus and its predator Thanasimus formicarius to bark beetle pheromones and host tree volatiles. J Comp Physiol A. 157:335–342. [Google Scholar]
- Tømmerås BÅ, Mustaparta H, Gregoire JC.. 1984. Receptor cells in Ips typographus and Dendroctonus micans specific to pheromones of the reciprocal genus. J Chem Ecol. 10(5):759–770. [DOI] [PubMed] [Google Scholar]
- Unelius RC, Schiebe C, Bohman B, Andersson MN, Schlyter F.. 2014. Non-host volatile blend optimization for forest protection against the European spruce bark beetle, Ips typographus. PLoS One. 9(1):e85381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Carey AF, Carlson JR, Zwiebel LJ.. 2010. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 107(9):4418–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Yi J, Li Y, Liu J, Wang J, Xi J.. 2020. Three host plant volatiles, hexanal, lauric acid, and tetradecane, are detected by an antenna-biased expressed odorant receptor 27 in the dark black chafer Holotrichia parallela. J Agric Food Chem. 68(28):7316–7323. [DOI] [PubMed] [Google Scholar]
- Wyatt TD. 2014. Pheromones and animal behavior: chemical signals and signatures. 2nd ed. Cambridge: Cambridge University Press. [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13(5):555–556. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 46(4):409–418. [DOI] [PubMed] [Google Scholar]
- Yuvaraj JK, Andersson MN, Corcoran JA, Anderbrant O, Löfstedt C.. 2018. Functional characterization of odorant receptors from Lampronia capitella suggests a non-ditrysian origin of the lepidopteran pheromone receptor clade. Insect Biochem Mol Biol. 100:39–47. [DOI] [PubMed] [Google Scholar]
- Yuvaraj JK, Corcoran JA, Andersson MN, Newcomb RD, Anderbrant O, Löfstedt C.. 2017. Characterization of odorant receptors from a non-ditrysian moth, Eriocrania semipurpurella sheds light on the origin of sex pheromone receptors in Lepidoptera. Mol Biol Evol. 34(11):2733–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj JK, Roberts RE, Sonntag Y, Hou X-Q, Grosse-Wilde E, Machara A, Zhang D-D, Hansson BS, Johanson U, Löfstedt C, et al. 2021. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 19(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-D, Löfstedt C.. 2013. Functional evolution of a multigene family: orthologous and paralogous pheromone receptor genes in the turnip moth, Agrotis segetum. PLoS One. 8(10):e77345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-D, Löfstedt C.. 2015. Moth pheromone receptors: gene sequences, function and evolution. Front Ecol Evol. 3:105. doi: 10.3389/fevo.2015.00105. [Google Scholar]
- Zhang D-D, Wang H-L, Schultze A, Froß H, Francke W, Krieger J, Löfstedt C.. 2016. Receptor for detection of a Type II sex pheromone in the winter moth. Sci Rep. 6:18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-D, Zhu KY, Wang CZ.. 2010. Sequencing and characterization of six cDNAs putatively encoding three pairs of pheromone receptors in two sibling species, Helicoverpa armigera and Helicoverpa assulta. J Insect Physiol. 56(6):586–593. [DOI] [PubMed] [Google Scholar]
- Zhang Q-H, Schlyter F.. 2004. Olfactory recognition and behavioural avoidance of angiosperm non-host volatiles by conifer bark beetles. Agric Forest Ent. 6(1):1–19. [Google Scholar]
- Zhao T, Ganji S, Schiebe C, Bohman B, Weinstein P, Krokene P, Borg-Karlson A-K, Unelius CR.. 2019. Convergent evolution of semiochemicals across Kingdoms: bark beetles and their fungal symbionts. Isme J. 13(6):1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Krokene P, Björklund N, Långström B, Solheim H, Christiansen E, Borg-Karlson A-K.. 2010. The influence of Ceratocystis polonica inoculation and methyl jasmonate application on terpene chemistry of Norway spruce, Picea abies. Phytochemistry 71(11–12):1332–1341. [DOI] [PubMed] [Google Scholar]
- Økland B, Bjørnstad ON.. 2003. Synchrony and geographical variation of the spruce bark beetle (Ips typographus) during a non-epidemic period. Popul Ecol. 45(3):213–219. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the oocyte recordings are presented in the main figures of the article. Sequences of the characterized ItypORs have been deposited in GenBank (accession numbers: MW556722–MW556726).