Abstract
Background
We previously reported that expression of a miR-138 mimic or knockdown of SIN3A in primary cultures of cystic fibrosis (CF) airway epithelia increased ΔF508-CFTR mRNA and protein levels, and partially restored CFTR-dependent chloride transport. Global mRNA transcript profiling in ΔF508-CFBE cells treated with miR-138 mimic or SIN3A siRNA identified two genes, SYVN1 and NEDD8, whose inhibition significantly increased ΔF508-CFTR trafficking, maturation, and function. Little is known regarding the dynamic changes in the CFTR gene network during such rescue events. We hypothesized that analysis of condition-specific gene networks from transcriptomic data characterizing ΔF508-CFTR rescue could help identify dynamic gene modules associated with CFTR biogenesis.
Methods
We applied a computational method, termed M-module, to analyze multiple gene networks, each of which exhibited differential activity compared to a baseline condition. In doing so, we identified both unique and shared gene pathways across multiple differential networks. To construct differential networks, gene expression data from CFBE cells were divided into three groups: (1) siRNA inhibition of NEDD8 and SYVN1; (2) miR-138 mimic and SIN3A siRNA; and (3) temperature (27 °C for 24 h, 40 °C for 24 h, and 27 °C for 24 h followed by 40 °C for 24 h).
Results
Interrogation of individual networks (e.g., NEDD8/SYVN1 network), combinations of two networks (e.g., NEDD8/SYVN1 + temperature networks), and all three networks yielded sets of 1-modules, 2-modules, and 3-modules, respectively. Gene ontology analysis revealed significant enrichment of dynamic modules in pathways including translation, protein metabolic/catabolic processes, protein complex assembly, and endocytosis. Candidate CFTR effectors identified in the analysis included CHURC1, GZF1, and RPL15, and siRNA-mediated knockdown of these genes partially restored CFTR-dependent transepithelial chloride current to ΔF508-CFBE cells.
Conclusions
The ability of the M-module to identify dynamic modules involved in ΔF508 rescue provides a novel approach for studying CFTR biogenesis and identifying candidate suppressors of ΔF508.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-021-01106-7.
Keywords: Cystic fibrosis, CFTR, Transcriptional profiling, Network biology, Gene modules, Gene ontology, M-module
Introduction
Cystic fibrosis (CF) is the most common lethal autosomal disease in Caucasian populations, affecting approximately 75,000 individuals worldwide [1, 2]. CF harms multiple organ systems and can cause meconium ileus, growth failure, diabetes, increased sweat chloride concentrations, and infertility, among other symptoms [3–10]. However, the majority of CF-associated morbidity and mortality results from chronic and progressive lung dysfunction, characterized by acidic airway surface liquid, weakened antimicrobial defenses at the airway surface, and impaired mucociliary transport leading to chronic bacterial airway infections, irreversible tissue remodeling, and respiratory failure.
CF is caused by mutations in cystic fibrosis transmembrane conductance regulator (CFTR) which encodes an anion channel. Although over 2000 mutations have been identified in CFTR, over 70% of disease-associated alleles contain a deletion of phenylalanine at position 508, termed ΔF508, correlating to roughly 90% of individuals with CF having one or two ΔF508 alleles [11–15]. The ΔF508-CFTR mutation results in protein misfolding and proteasomal degradation [16, 17]. The observation that low temperature (27 °C) incubation could rescue and traffic ΔF508-CFTR to the cell surface was compelling because it demonstrated that partial function could be retained if ΔF508-CFTR escaped the endoplasmic reticulum-associated degradation (ERAD) pathway and trafficked to the cell membrane [16]. Although manipulation of temperature is not therapeutically practical, this observation encouraged the investigation of genes affecting the processing and maturation of CFTR. We previously identified microRNA-138, SIN3A, NEDD8, and SYVN1 as members of the CFTR biogenesis pathway [18, 19]. Functional assays determined that miR-138 overexpression or siRNA knockdown of SIN3A, SYVN1, or NEDD8 partially restored the maturation, trafficking, and function of ΔF508-CFTR.
Additional transcriptomic studies have identified effectors of CFTR biogenesis. For example, Clarke and colleagues performed a microarray study of primary epithelial cells from ΔF508 homozygotes and non-CF controls, which yielded a molecular signature of native CF airway epithelial cells in which a noteworthy number of genes involved in inflammation and defense were upregulated, including S100A8, S100A9, and SERPINA3 [20]. A follow-up meta-analysis by Clark et al. identified several negative regulators of CFTR, including SNX6, PSEN1, and RCN2 [21]. When these genes were knocked down via siRNA, a considerable increase in CFTR trafficking to the cell membrane was observed.
Despite the identification of several in vitro effectors of ΔF508-CFTR correction, its biogenesis pathway remains incompletely understood. Analyzing and understanding rescue-specific molecular events are critical for understanding CF biogenesis and the development of therapeutics. Network biology is a powerful tool for analyzing such complex disease phenotypes. For example, Taylor et al. demonstrated that hub gene topology could be used to improve the prognosis of breast cancer, while Chuang and colleagues revealed that differentially expressed subnetworks are effective biomarkers for breast cancer metastasis [22, 23]. However, a common theme of network biology is the dichotomization of disease progression, either for the onset or severity of disease, as many studies analyze each condition individually. Such studies highlight hubs, modules, or edges that are significantly associated with only a single condition, rather than modeling the gene expression data as a single continuum. This ultimately limits our ability to observe changes at a pathway level during disease progression.
Likewise, focusing on only CF vs. non-CF omics data, or the analysis of only a single rescue signature, may limit the detection of CFTR biogenesis interactors. To address this critical gap in network biology, Tan and colleagues developed a general framework, termed M-module, to reveal subnetwork dynamics by joint analysis of multiple gene co-expression networks [24, 25]. They demonstrated that the use of network connectivity dynamics significantly improved the classification accuracy of multiple breast cancer stages. Here, we use this M-module framework to analyze multiple gene co-expression networks relevant to CFTR to identify interactions favoring CFTR biogenesis (Table 1). These networks include miR-138 overexpression and SIN3A knockdown, NEDD8 and SYVN1 knockdown, and low temperature treatments.
Table 1.
Co-expression networks analyzed using the M-module framework
| Group | Control | Treatments |
|---|---|---|
| Temperature | 37 °C | 27 °C for 24 h, 40 °C for 24 h, 27 °C for 24 h followed by 40 °C for 24 h |
| NEDD8 | Scrambled siRNA | siRNA inhibition of NEDD8 and SYVN1 |
| miR-138 | Scrambled siRNA | miR-138 mimic and siRNA inhibition of SIN3A |
Materials and methods
M-module analysis
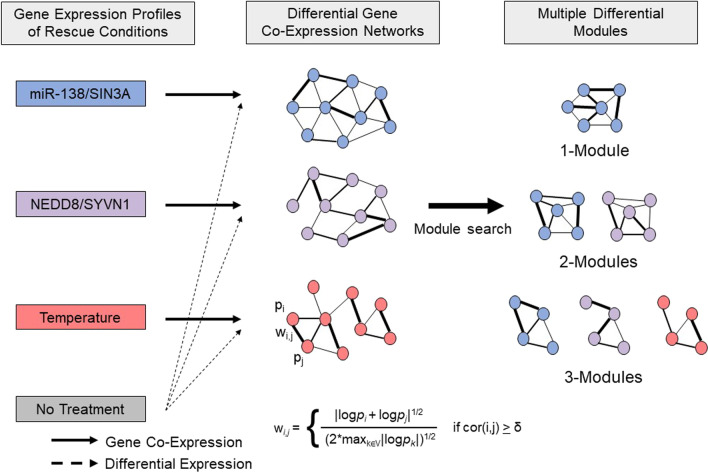
To identify both unique and shared gene pathways across multiple differential networks, we grouped gene expression data from CFBE41o− cells (GSE142610) into three conditions (Table 1). The first group, termed “miR-138/SIN3A”, compared overexpression of miR-138 and siRNA knockdown of SIN3A with a scrambled siRNA control [18]. The second group, termed “NEDD8/SYVN1”, compared siRNA knockdown of NEDD8 and SYVN1 compared to a scrambled siRNA control [19]. The third group, termed “Temperature”, included treatment conditions of 27 °C for 24 h, 40 °C for 24 h, and 27 °C for 24 h followed by 40 °C for 24 h, compared to a 37 °C control [16]. A summary and full lists of differentially expressed genes across conditions can be found in Additional file 1: Tables S1-S4, and Figure S1. We used a literature-based curated list of 333 CFTR-associated genes, termed the “CFTR Interactome”, as seed nodes (Additional file 1: Table S5). The gene expression profiles across the multiple rescue conditions were superimposed on human protein–protein interaction networks and used to build differential gene networks [26–29]. First, we constructed a binary expression network with edges chosen based on the absolute value of Pearson correlation of the expression profiles of two genes (i,j). Edges whose correlation did not exceed a pre-defined threshold δ were removed from the binary network. Remaining edges were then weighted (wi,j) using the geometric mean of the p values (pi and pj) of differential gene expression between the baseline and rescue conditions. Second, multiple differential networks were analyzed to identify multiple differential modules (M-DMs) under different conditions. 1-Modules are modules that were only found in one experimental condition whereas M-Modules with M ≥ 2 are modules that were found in multiple conditions.
Module gene significance
Module gene significance was determined by running 10,000 randomized iterations of the M-module analysis and calculating average distances of non-seed genes to seed genes. If the distance was significantly less in the randomized networks, then the gene was considered insignificant in our module output. Genes with FDR < 0.01 were considered significant.
Gene ontology enrichment analysis
We next ranked non-seed significant module genes by minimum distance to closest seed genes. Gene Ontology (GO) enrichment analysis was performed on the non-seed significant module genes. For each enriched GO term, the average minimum distance between contributing non-seed module genes and CFTR-associated seed genes was calculated. Such analyses allowed for the unbiased identification of candidate genes potentially involved in CFTR-relevant pathways and functions.
Cultured cells
CFBE41o− cells, termed “CFBE cells”, originally developed by immortalization of CF airway epithelial cells and later transduced with a ΔF508-CFTR expression cassette using the TranzVector lentivirus system were cultured as previously described [30–32]. These cells were obtained from Dr. J.P. Clancy at Cincinnati Children’s Hospital.
Oligonucleotide reagents
Dicer-Substrate Short Interfering RNAs (DsiRNAs) were obtained as TriFECTa kits from IDT (Coralville, IA), each containing three pre-designed siRNAs per gene. Transfections were performed in 24-well plates, with 3 μL of Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) and 1 μL of each 10 μM DsiRNA added to 94 μL of Opti-MEM (Gibco, Waltham, MA) media per well. Following a five minute incubation at room temperature, 50 μL of the DsiRNA-lipid complex was added to each well and 200s cells were suspended in 100 μL of Opti-MEM and seeded onto 24-well plates. Knockdown efficiency was measured following 24 h of incubation at 37 °C. To ascertain the specificity of the oligonucleotides, we harvested RNA from cells transfected with the three pooled oligonucleotides per gene and measured the expression of multiple genes 24 h post-transfection. First-strand cDNA was synthesized using SuperScript II (Invitrogen, Carlsbad, CA) with oligo-dT and random-hexamer primers. Primers for each gene were designed and produced by IDT and validated in HEK cells. Quantitative RT-PCR was performed using the QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems, Foster City, CA). All experiments were performed in quadruplicate. Following validation of knockdown efficiency, three siRNAs per gene were pooled and CFBE cells were reverse-transfected and grown on microporous membranes of Transwell (Corning, Corning, NY) plates seven days prior to the electrophysiology measurements [33]. Oligo sequences and siRNA knockdown efficiencies can be found in Additional file 1: Table S6.
Transepithelial chloride current studies
Transepithelial chloride current measurements were made in Ussing chambers approximately seven days post-seeding. Briefly, epithelial sheets were mounted in the Ussing chamber and transepithelial chloride current (It) was measured under short-circuit conditions. After measuring baseline current, 100 μM amiloride (Amil) was added apically to inhibit epithelial sodium channels (ENaC) followed by apical addition of 100 μM 4,4′-diisothiocyanoto-stilbene-2,2′-disulfonic acid (DIDS) to inhibit non-CFTR chloride channels. Next, we applied 10 μM forskolin and 100 μM 3-isobutyl-1-methylxanthine (IBMX). These agents elevate intracellular cAMP levels leading to the PKA-mediated phosphorylation and activation of CFTR channels. Finally, 100 μM GlyH-101, an inhibitor of CFTR channels, was added apically. Studies were conducted with 135 mM NaCl, 1.2 mM MgCl2, 1.2 mM CaCl2, 2.4 mM K2PO4, 0.6 mM KH2PO4, 5 mM dextrose, and 5 mM HEPES (pH 7.4) on the basolateral surface, and an apical chloride concentration gradient with gluconate substituted for chloride.
Statistical analysis
For electrophysiology studies, the average change in peak transepithelial current (It) was calculated and statistical significance was determined using the Brown-Forsythe ANOVA with Benjamini–Hochberg multiple comparison correction (*p < 0.05) [34]. Data are presented as a mean ± standard error of individual data points. p < 0.05 was considered significant.
Results
Identification of individual rescue condition-specific modules and networks
As shown in Fig. 1, the miR-138, NEDD8, and temperature co-expression profiles were used to create differential networks that yielded multiple M-Modules. Static 1-Modules were unique to an individual rescue condition, where 2-Modules were modules found across two networks and 3-Modules were modules found across all networks. As shown in Table 2, 70 1-Modules containing 964 significant genes were identified in the miR-138 network. The NEDD8 network yielded 55 1-Modules containing 764 significant genes. 44 1-Modules containing 342 significant genes were found in the temperature network. The miR-138 network shared 13 2-Modules containing 159 significant genes with the NEDD8 network and 19 2-Modules containing 191 significant genes with the temperature network. The NEDD8 and temperature networks shared 10 2-Modules containing 160 significant genes. All rescue conditions shared seven 3-Modules containing 103 significant genes.
Fig. 1.
Rescue conditions queried in the M-module framework. Gene expression profiles across the miR-138/SIN3A, NEDD8/SYVN1, and Temperature rescue conditions used to build differential gene co-expression networks. A binary co-expression network was constructed in which genes were selected based on the absolute value of Pearson correlation of the expression profiles of two genes. Only edges whose correlation exceeded a pre-defined threshold were included. Edges were then weighted (wi,j) based on the p values (pi and pj) of differential gene expression between the baseline and rescue conditions. Multiple differential co-expression networks were then analyzed to identify shared and unique multiple differential modules. 1-Modules are unique to a single rescue condition, whereas 2-Modules are found across two rescue conditions, and 3-Modules are found in all three rescue conditions. Adapted from [24]
Table 2.
M-module query results
| Type of module | Condition | # of modules | # of unique genes | # of significant genes (FDR < 0.01) |
|---|---|---|---|---|
| 1-DM | miR-138 | 70 | 3109 | 964 |
| NEDD8 | 55 | 2438 | 764 | |
| Temperature | 44 | 1433 | 342 | |
| 2-DM | miR-138 + NEDD8 | 13 | 393 | 159 |
| miR-138 + Temperature | 19 | 654 | 191 | |
| NEDD8 + Temperature | 10 | 305 | 160 | |
| 3-DM | miR-138 + NEDD8 + Temperature | 7 | 311 | 103 |
Gene ontology analysis of non-seed module genes identifies enriched CF-relevant terms
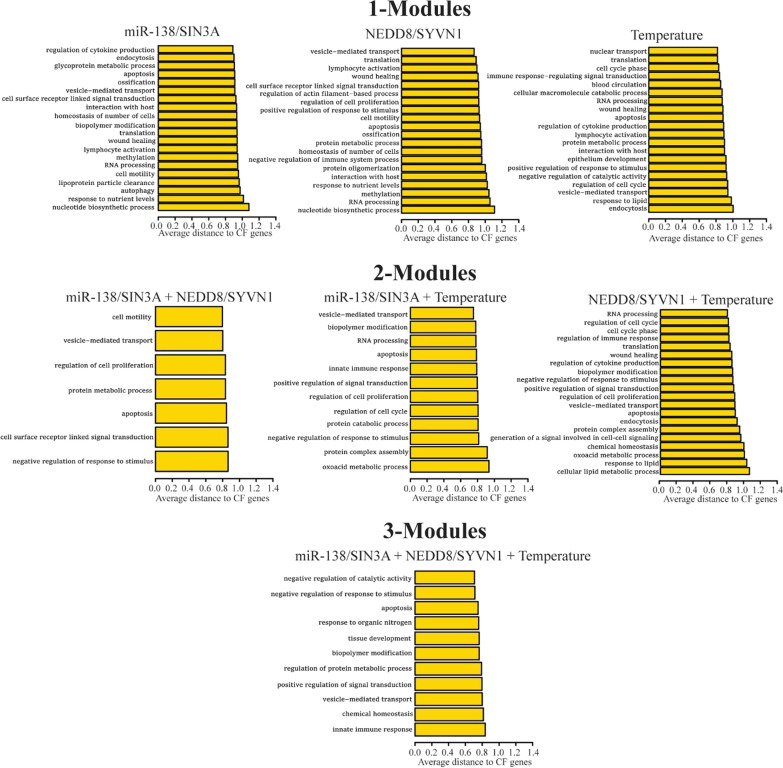
We ranked non-seed significant module genes by minimum distance (the sum of weight of the shortest path between two nodes) to closest seed genes. Gene Ontology (GO) enrichment analysis was performed on the non-seed significant module genes. For each enriched GO term, the average minimum distance between contributing non-seed module genes and CFTR-associated seed genes was calculated. Several CFTR-relevant pathways were identified using this unbiased approach, further validating that M-module could connect genes previously unknown to affect CFTR biogenesis to ΔF508-CFTR-related functions. For example, the following terms were enriched in at least two conditions: translation, protein metabolic/catabolic processes, protein complex assembly, endocytosis, vesicle-mediated transport, apoptosis, and autophagy, among others (Fig. 2).
Fig. 2.
Gene Ontology analysis of non-seed module genes. Enriched terms for significant non-seed module genes found exclusively in 1-Differential Modules (DMs) (top panel), 2-DMs (middle panel), or 3-DMs (bottom panel). X-axis represents the average distance of significant non-seed module genes to CF seed genes
Identification of candidate CFTR effectors
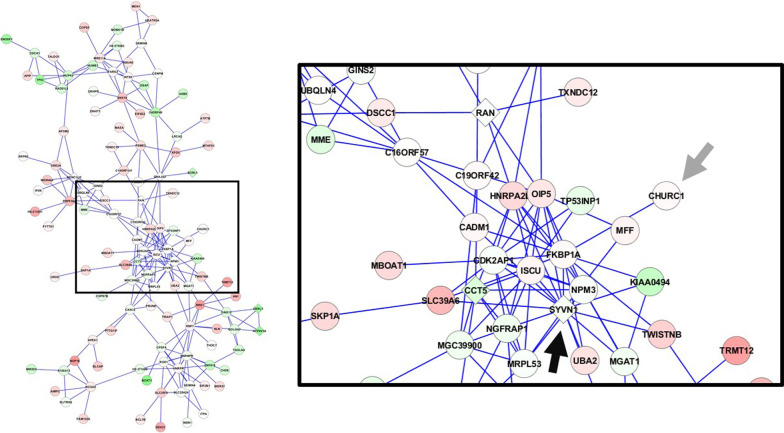
To identify novel CFTR effectors, significance was first determined by comparing individual module genes to a randomized network. Genes with a FDR < 0.01 were considered significant, narrowing the number of genes across all modules and conditions from 8643 to 2683. Next, all CFTR interactome genes were removed, as were all genes that had previously been tested in various studies, leaving 901 untested non-seed module genes (Additional file 1: Table S7) [18, 19, 35–47]. Due to the known interactions between CFTR and several E3 ubiquitin ligases, we chose to investigate ASB6 and ASB13 (components of the SOCS-box ubiquitin ligase complex), FBXO46 (F-box), ZFAND5 (zf-A20), and the BTB domain-containing proteins GZF1, KLHL29, and ZBTB38 [19, 36, 37, 48–62]. As Lukacs and colleagues recently identified several components of the ribosomal stalk as being CFTR effectors, we also elected to study RPL15, RPL28, and RPL39L [63]. Lastly, we tested non-seed module genes whose closest seed neighbor was the ERAD-associated protein SYVN1, as our previous studies demonstrated that SYVN1 knockdown restored partial function to ΔF508-CFTR [19]. These genes included C11ORF1, CADM1, CHURC1, JPT1, MFF, POLR1F, RRS1, and THOC7. Figure 3 displays a 1-Module containing CHURC1 and the neighboring SYVN1.
Fig. 3.
Example 1-Module containing CHURC1. The miR-138/SIN3A differential gene co-expression network yielded 70 unique 1-modules, including the 1-Module shown here. Diamonds represent seed genes, while circles signify non-seed genes. The node color is proportional to the −log10 p value of gene expression, with red indicating up-regulation and green down-regulation. The inset contains CHURC1 (gray arrow) and shows that its nearest seed neighbor is SYVN1 (black arrow). This figure was created using Cytoscape [83]
siRNA-mediated knockdown of CHURC1, GZF1, and RPL15 restores partial function to ΔF508-CFTR
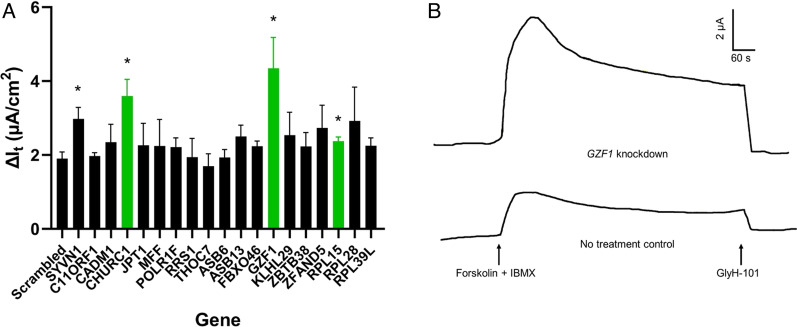
We used pools of three siRNAs per gene to knockdown candidate gene mRNA transcripts in CFBE cells, using the change in cAMP-activated chloride secretion as an endpoint [33]. Knockdown of CHURC1 significantly improved CFTR-dependent transepithelial chloride current in ΔF508-CFTR CFBE cells, with an increase of 89% in peak chloride current after the addition of forskolin and IBMX compared to the scrambled control (p = 0.0112; Fig. 4). Compared to knockdown of SYVN1, CHURCH1 knockdown increased peak chloride current by over 20%. Knockdown of GZF1 resulted in an increase of 129% in peak chloride current when compared to the scrambled control (p = 0.0405) and an increase of 46% compared to SYVN1. Lastly, knockdown of RPL15 also produced a statistically significant ~ 25% increase in cAMP-activated, GlyH-101-sensitive transepithelial chloride current compared to the scrambled control (p = 0.0446). Representative transepithelial current tracings showing CFTR-dependent chloride current in CFBE cells treated with a scrambled control or siRNA targeting GZF1 are shown in Fig. 4b. Additional representative tracings of knockdown of CHURCH1, RPL15, and THOC7 are displayed in Additional file 1: Fig. S2.
Fig. 4.
siRNA-mediated knockdown of CHURC1, GZF1, and RPL15 rescues ΔF508-CFTR function in CFBE cells. a Average change in transepithelial current (It) in response to the cAMP agonists forskolin and IBMX (F&I) and the CFTR inhibitor GlyH-101 under open circuit conditions was measured in CFBE cells. Three siRNAs per gene were pooled and CFBE cells were reverse-transfected using Lipofectamine RNAiMAX and grown on microporous Transwell membranes seven days prior to the electrophysiology measurements. Error bars indicate standard error. Statistical significance compared to the scrambled control was determined by Brown-Forsythe ANOVA and post-hoc Benjamini–Hochberg multiple testing correction (*p < 0.05). n = 4–6 per gene. CHURC1, GZF1, and RPL15 are highlighted in green. b Representative transepithelial current tracings showing CFTR-dependent chloride current in CFBE cells treated with a scrambled control or siRNA targeting GZF1. The Y-axis represents transepithelial current in µA and the X-axis represents time in seconds
Discussion
While several transcriptomics-based studies have identified effectors, CFTR biogenesis remains incompletely understood. The analysis of the underlying molecular events responsible for ΔF508-CFTR rescue is crucial for understanding the CFTR biosynthetic process and the development of therapeutics. As many transcriptomic and network biology studies of CF focus on diseased versus healthy cells, the resulting seed genes, subnetworks, and edge sets are significantly associated with only one condition and this may limit the detection of CFTR interactors. To address critical gaps in the knowledge of CFTR biogenesis, we applied the M-module framework to reveal subnetwork dynamics by joint analysis of multiple co-expression networks representing ΔF508-CFTR rescue. These networks included miR-138 overexpression and SIN3A knockdown, NEDD8 and SYVN1 knockdown, and low temperature treatments.
As displayed in Table 2, dozens of unique static modules were identified in the individual conditions, as well as many dynamic modules that were shared between two or all conditions. To assess the quality of these modules, we performed gene ontology (GO) analyses on all significant non-seed module genes across all conditions. Removing all CF-related genes allowed for an unbiased assessment of the functional gene groups represented by the modules. Rather than CF-related genes pushing the gene ontology analysis toward enrichment of CF-related terms, we could instead assess GO enrichments driven exclusively by genes previously unassociated with CFTR. As shown in Fig. 2, many GO enriched terms were related to CF and multiple terms were present across several conditions. For example, genes involved in translation, protein metabolic/catabolic processes, and protein complex assembly were enriched in multiple conditions and both 1- and 2-Modules. As ΔF508-CFTR is classified as a protein folding defect, it is perhaps not surprising that signatures of its rescue were highly enriched for genes involved in translation and protein metabolism [16, 17]. However, the removal of all CFTR-related genes prior to gene ontology analysis means that only genes previously unlinked to CFTR biogenesis were responsible for such GO enrichments. Therefore, is it likely that our modules contain previously undiscovered CFTR effectors that contribute to ΔF508-CFTR rescue.
Likewise, GO analysis indicated that our non-seed modules were enriched for genes involved in endocytosis, vesicle-mediated transport, autophagy, and apoptosis. CFTR is regulated in part by cAMP-dependent vesicle traffic to the apical membrane, although ΔF508-CFTR is typically degraded before reaching the cell surface [64]. Even if mutant protein reaches the cell membrane, clathrin-mediated endocytosis efficiently removes it from the cell surface via recycling endosomes and the protein is directed to lysosomes for degradation [65, 66]. Furthermore, ΔF508-CFTR causes dysfunction of apoptotic and autophagic processes, which may contribute to CF lung disease progression [67–69]. These gene ontology results strongly support the likelihood of our non-seed module genes being members of the CFTR interactome and contributing to its biogenesis.
As our gene ontology analyses suggested that non-seed module genes may contribute to ΔF508-CFTR rescue, we aimed to identify specific genes with such functions. By focusing on genes with SYVN1 as their nearest seed neighbor, we hypothesized that CHURC1, among several other genes, could affect CFTR rescue. siRNA-mediated knockdown of CHURC1 rescued CFTR-dependent transepithelial chloride current to ΔF508-CFTR by 89% compared to a scrambled control. Knockdown of CHURC1 even produced an increase of over 20% in peak chloride current when compared to knockdown of SYVN1. CHURC1 (Churchill Domain Containing 1) is a transcriptional activator that mediates fibroblast growth factor (FGF) signaling during neural development in zebrafish [70–73]. Rotin and colleagues recently identified FGF Receptor 1 (FGFR1) as a suppressor of ΔF508-CFTR maturation [46]. Further analysis revealed that knockdown of FGFR1, FGFR2, FGFR3, and downstream signaling proteins ERK1, ERK2, AKT, PLCγ-1, and FRS2α, increased ΔF508-CFTR channel activity and protein maturation. These data suggest that CHURC1 knockdown may rescue mutant CFTR function through the FGF signaling pathway.
To further elucidate potential mechanisms of CHURC1, we queried the gene in ARChS4, which allows for massive mining of publicly available RNA-seq data [74]. Interestingly, the highest ranked predicted GO biological process for CHURC1 was ribosomal small subunit biogenesis (Additional file 1: Table S8). Additional enriched terms included cotranslational protein targeting to membrane, protein targeting to ER, translational elongation, ribosomal large subunit biogenesis, translational initiation, and protein complex assembly. Lukacs and colleagues recently identified a component of the ribosomal stalk, RPL12, as an effector of ΔF508-CFTR [63]. Silencing of RPL12 slowed the rate of translation, while increasing the folding efficiency and conformational stability of ΔF508-CFTR. Furthermore, RPL12 knockdown in combination with lumacaftor restored ΔF508-CFTR function to approximately 50% of the wild-type channel in primary human airway epithelial cells. Lukacs and colleagues also observed that silencing of ribosome stalk proteins RPLP0, RPLP1, and RPLP2 partially rescued ΔF508-CFTR function. In our siRNA knockdown screen, knockdown of RPL15, also a protein member of the ribosomal stalk, significantly restored CFTR-dependent chloride secretion. Similar to CHURC1, the GO analysis results for RPL15 were enriched for ribosome-related processes (Additional file 1: Table S9). While CHURC1 was identified in all three 1-Module conditions, RPL15 was identified in the NEDD8 and Temperature 1-Modules. Our M-module analysis not only identified CHURC1 and RPL15 as potential effectors of CFTR rescue, it also suggests that an underlying mechanism of our queried rescue signatures is the manipulation of translation and the ribosome.
Lending additional support to the candidacy of CHURC1 as a CFTR effector is the Encyclopedia of DNA Elements (ENCODE), which provides evidence for SIN3A transcription factor binding at the promotor of CHURC1 in several cell types, including the A549 respiratory epithelial cell line [75, 76]. SIN3A can also bind the promoter of RPL15 in A549 cells, according to ENCODE. Furthermore, TargetScan predicts that miR-138 regulates expression of CHURC1 [77]. Therefore, it is also possible that CHURC1 and RPL15 act through the miR-138/SIN3A pathway we previously described [18]. While GO analysis of GZF1 did not yield ribosomal- or CFTR-related biological processes (Additional file 1: Table S10), ENCODE and ChIP Enrichment Analysis (ChEA) Transcription Factor Targets both indicate that FOXA1 and FOXA2 bind the promoter of GZF1 [78, 79]. Harris and colleagues demonstrated that depletion of FOXA1 and FOXA2 represses CFTR expression. Interestingly, these forkhead box transcription factors have also been shown to act as transcriptional repressors [80, 81]. GZF1 was identified in a miR-138/SIN3A 1-Module and ENCODE also suggests that SIN3A binds the promoter of GZF1, supporting the hypothesis that GZF1 may also act through the miR-138/SIN3A pathway.
Conclusion
The ability of the M-module to identify dynamic modules involved in ΔF508-CFTR rescue provided a novel approach for studying CFTR biogenesis and allowed for the identification of previously unknown CFTR effectors. siRNA-mediated knockdown of CHURC1, GZF1, and RPL15 significantly restored CFTR-dependent transepithelial chloride current in ΔF508-CFTR CFBE cells. Further analysis of these genes and their roles in ΔF508-CFTR rescue and CFTR biogenesis will be required to elucidate exact mechanisms of action.
Supplementary Information
Additional file 1. Table S1. Summary of differentially expressed genes across conditions in CFBE cells. Summary of up- and down-regulated differentially expressed genes in conditions vs. controls. Table S2. Differentially expressed genes in the miR-138/SIN3A condition. List of differentially expressed genes in the miR-138/SIN3A conditions vs. scrambled siRNA control. Table S3. Differentially expressed genes in the NEDD8/SYVN1 condition. List of differentially expressed genes in the NEDD8/SYVN1 conditions vs. scrambled siRNA control. Table S4. Differentially expressed genes in the temperature condition. List of differentially expressed genes in the temperature conditions vs. 37°C control. Table S5. CFTR interactome used as seed nodes. List of CFTR effectors and interactors used as seed nodes in the M-module analysis. Table S6. DsiRNA and primer sequences. List of siRNA and primer sequences used in the functional knockdown experiments to test for CFTR rescue. Table S7. Untested non-seed module genes. List of genes resulting from the M-module analysis that have not been previously tested or linked to CFTR. Table S8. Top 50 predicted gene ontology biological processes for CHURC1. List of biological processes associated with CHURC1 according to the ARChS4 software. Table S9. Top 50 predicted gene ontology biological processes for RPL15. List of biological processes associated with RPL15 according to the ARChS4 software. Table S10. Top 50 predicted gene ontology biological processes for GZF1. List of biological processes associated with GZF1 according to the ARChS4 software. Figure S1. Schematic showing intersection of differentially expressed genes across conditions. Controls and conditions are described in Table 1. Up arrows indicate up-regulated genes; down arrows indicate down-regulated genes. Significance is defined as FDR < 0.05. Figure S2. Representative transepithelial current tracings demonstrating the effects of individual gene knockdown on CFTR-dependent chloride current in CFBE cells. The Y-axis represents transepithelial current in µA and the X-axis represents time in seconds. The addition of the cAMP agonists forskolin and IBMX resulted in an increase in CFTR-dependent transepithelial chloride current in cells treated with DsiRNAs targeting: A) CHURC1 or B) RPL15. This increase in current was inhibited by the CFTR channel inhibitor GlyH-101. The tracing shown in C demonstrates that DsiRNA knockdown of THOC7 was ineffective in restoring CFTR-dependent chloride current.
Acknowledgements
We thank Drs. Jennifer Bartlett, Miguel Ortiz, and Andrew Thurman for their critical review of the manuscript. An early version of this study was presented in abstract form at the 29th North American Cystic Fibrosis Conference, October 8-10, 2015.
Abbreviations
- CF
Cystic fibrosis
- CFBE
Cystic fibrosis bronchial epithelia
- CFTR
Cystic fibrosis transmembrane conductance regulator
- DIDS
4,4′-Diisothiocyanoto-stilbene-2,2′-disulfonic acid
- DsiRNA
Dicer-substrate short interfering RNA
- ERAD
Endoplasmic reticulum-associated degradation
- GO
Gene ontology
- IBMX
3-Isobutyl-1-methylxanthine
Authors’ contributions
M.D.S., L.G., K.T., and P.B.M. designed experiments. M.D.S. and L.G. performed experiments. M.D.S., L.G., K.T., and P.B.M. analyzed and interpreted data. M.D.S. and P.B.M. were the primary contributors in writing the manuscript, and L.G. and K.T. provided feedback. All authors read and approved the final manuscript.
Funding
This work was supported by National Institutes of Health (NIH) Grant R01 HL118000 (PBM) and Cystic Fibrosis Foundation (PBM), MDS is supported by NIH Fellowship 5T32 GM008629 (Daniel Eberl). The authors also acknowledge the support of the In Vitro Models and Cell Culture Core, partially supported by NIH Grant P30 DK054759 and the Cystic Fibrosis Foundation. PBM is supported by the Roy J. Carver Charitable Trust. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets analyzed during the current study are available in the Gene Expression Omnibus Series GSE142610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142610) [82]. M-module software can be installed at https://github.com/tanlabcode/M-module. Individual modules and datasets are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ratjen F, Doring G. Cystic fibrosis. Lancet. 2003;361(9358):681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 2.Brown SD, White R, Tobin P. Keep them breathing: cystic fibrosis pathophysiology, diagnosis, and treatment. JAAPA. 2017;30(5):23–27. doi: 10.1097/01.JAA.0000515540.36581.92. [DOI] [PubMed] [Google Scholar]
- 3.Fakhoury K, Durie PR, Levison H, Canny GJ. Meconium ileus in the absence of cystic fibrosis. Arch Dis Child. 1992;67(10 Spec No):1204–1206. doi: 10.1136/adc.67.10_Spec_No.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly A, Moran A. Update on cystic fibrosis-related diabetes. J Cyst Fibros. 2013;12(4):318–331. doi: 10.1016/j.jcf.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Kobelska-Dubiel N, Klincewicz B, Cichy W. Liver disease in cystic fibrosis. Prz Gastroenterol. 2014;9(3):136–141. doi: 10.5114/pg.2014.43574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quinton PM. Missing Cl conductance in cystic fibrosis. Am J Physiol. 1986;251(4 Pt 1):C649–C652. doi: 10.1152/ajpcell.1986.251.4.C649. [DOI] [PubMed] [Google Scholar]
- 7.Sokol RZ. Infertility in men with cystic fibrosis. Curr Opin Pulm Med. 2001;7(6):421–426. doi: 10.1097/00063198-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Thalhammer GH, Eber E, Uranus S, Pfeifer J, Zach MS. Partial splenectomy in cystic fibrosis patients with hypersplenism. Arch Dis Child. 2003;88(2):143–146. doi: 10.1136/adc.88.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesdachai S, Tangpricha V. Treatment of vitamin D deficiency in cystic fibrosis. J Steroid Biochem Mol Biol. 2016;164:36–39. doi: 10.1016/j.jsbmb.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scaparrotta A, Di Pillo S, Attanasi M, Consilvio NP, Cingolani A, Rapino D, et al. Growth failure in children with cystic fibrosis. J Pediatr Endocrinol Metab. 2012;25(5-6):393–405. doi: 10.1515/jpem-2012-0012. [DOI] [PubMed] [Google Scholar]
- 11.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 12.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245(4922):1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 13.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245(4922):1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 14.Castellani C, team C. CFTR2: how will it help care? Paediatr Respir Rev. 2013;14(Suppl 1):2–5. [DOI] [PubMed]
- 15.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352(19):1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 16.Denning GM, Ostedgaard LS, Welsh MJ. Abnormal localization of cystic fibrosis transmembrane conductance regulator in primary cultures of cystic fibrosis airway epithelia. J Cell Biol. 1992;118(3):551–559. doi: 10.1083/jcb.118.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kartner N, Augustinas O, Jensen TJ, Naismith AL, Riordan JR. Mislocalization of delta F508 CFTR in cystic fibrosis sweat gland. Nat Genet. 1992;1(5):321–327. doi: 10.1038/ng0892-321. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran S, Karp PH, Jiang P, Ostedgaard LS, Walz AE, Fisher JT, et al. A microRNA network regulates expression and biosynthesis of wild-type and DeltaF508 mutant cystic fibrosis transmembrane conductance regulator. Proc Natl Acad Sci USA. 2012;109(33):13362–13367. doi: 10.1073/pnas.1210906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran S, Osterhaus SR, Parekh KR, Jacobi AM, Behlke MA, McCray PB., Jr SYVN1, NEDD8, and FBXO2 proteins regulate DeltaF508 cystic fibrosis transmembrane conductance regulator (CFTR) ubiquitin-mediated proteasomal degradation. J Biol Chem. 2016;291(49):25489–25504. doi: 10.1074/jbc.M116.754283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res. 2013;14:38. doi: 10.1186/1465-9921-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke LA, Botelho HM, Sousa L, Falcao AO, Amaral MD. Transcriptome meta-analysis reveals common differential and global gene expression profiles in cystic fibrosis and other respiratory disorders and identifies CFTR regulators. Genomics. 2015;106(5):268–277. doi: 10.1016/j.ygeno.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 24.Ma X, Gao L, Karamanlidis G, Gao P, Lee CF, Garcia-Menendez L, et al. Revealing pathway dynamics in heart diseases by analyzing multiple differential networks. PLoS Comput Biol. 2015;11(6):e1004332. doi: 10.1371/journal.pcbi.1004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Gao L, Tan K. Modeling disease progression using dynamics of pathway connectivity. Bioinformatics. 2014;30(16):2343–2350. doi: 10.1093/bioinformatics/btu298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47(D1):D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36(Datebase issue):D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kunzelmann K, Schwiebert EM, Zeitlin PL, Kuo WL, Stanton BA, Gruenert DC. An immortalized cystic fibrosis tracheal epithelial cell line homozygous for the delta F508 CFTR mutation. Am J Respir Cell Mol Biol. 1993;8(5):522–529. doi: 10.1165/ajrcmb/8.5.522. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Wakefield JK, Liu H, Xiao H, Kralovics R, Prchal JT, et al. Development of a novel trans-lentiviral vector that affords predictable safety. Mol Ther. 2000;2(1):47–55. doi: 10.1006/mthe.2000.0095. [DOI] [PubMed] [Google Scholar]
- 32.Bebok Z, Collawn JF, Wakefield J, Parker W, Li Y, Varga K, et al. Failure of cAMP agonists to activate rescued deltaF508 CFTR in CFBE41o- airway epithelial monolayers. J Physiol. 2005;569(Pt 2):601–615. doi: 10.1113/jphysiol.2005.096669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran S, Krishnamurthy S, Jacobi AM, Wohlford-Lenane C, Behlke MA, Davidson BL, et al. Efficient delivery of RNA interference oligonucleotides to polarized airway epithelia in vitro. Am J Physiol Lung Cell Mol Physiol. 2013;305(1):L23–32. doi: 10.1152/ajplung.00426.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57:289–300. [Google Scholar]
- 35.Bomberger JM, Barnaby RL, Stanton BA. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J Biol Chem. 2009;284(28):18778–18789. doi: 10.1074/jbc.M109.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caohuy H, Jozwik C, Pollard HB. Rescue of DeltaF508-CFTR by the SGK1/Nedd4-2 signaling pathway. J Biol Chem. 2009;284(37):25241–25253. doi: 10.1074/jbc.M109.035345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grove DE, Fan CY, Ren HY, Cyr DM. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRDeltaF508. Mol Biol Cell. 2011;22(3):301–314. doi: 10.1091/mbc.e10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harada K, Okiyoneda T, Hashimoto Y, Ueno K, Nakamura K, Yamahira K, et al. Calreticulin negatively regulates the cell surface expression of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2006;281(18):12841–12848. doi: 10.1074/jbc.M512975200. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127(4):803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- 40.Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329(5993):805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins LA, Fisher GW, Naganbabu M, Schmidt BF, Mun F, Bruchez MP. High-content surface and total expression siRNA kinase library screen with VX-809 treatment reveals kinase targets that enhance F508del-CFTR rescue. Mol Pharm. 2018;15(3):759–767. doi: 10.1021/acs.molpharmaceut.7b00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pankow S, Bamberger C, Calzolari D, Martinez-Bartolome S, Lavallee-Adam M, Balch WE, et al. F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528(7583):510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutt DM, Herman D, Rodrigues AP, Noel S, Pilewski JM, Matteson J, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6(1):25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomati V, Pesce E, Caci E, Sondo E, Scudieri P, Marini M, et al. High-throughput screening identifies FAU protein as a regulator of mutant cystic fibrosis transmembrane conductance regulator channel. J Biol Chem. 2018;293(4):1203–1217. doi: 10.1074/jbc.M117.816595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomati V, Sondo E, Armirotti A, Caci E, Pesce E, Marini M, et al. Genetic inhibition of the ubiquitin ligase Rnf5 attenuates phenotypes associated to F508del cystic fibrosis mutation. Sci Rep. 2015;5:12138. doi: 10.1038/srep12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trzcinska-Daneluti AM, Chen A, Nguyen L, Murchie R, Jiang C, Moffat J, et al. RNA interference screen to identify kinases that suppress rescue of DeltaF508-CFTR. Mol Cell Proteomics. 2015;14(6):1569–1583. doi: 10.1074/mcp.M114.046375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trzcinska-Daneluti AM, Ly D, Huynh L, Jiang C, Fladd C, Rotin D. High-content functional screen to identify proteins that correct F508del-CFTR function. Mol Cell Proteomics. 2009;8(4):780–790. doi: 10.1074/mcp.M800268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531(2):339–342. doi: 10.1016/S0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- 49.Alberti S, Bohse K, Arndt V, Schmitz A, Hohfeld J. The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator. Mol Biol Cell. 2004;15(9):4003–4010. doi: 10.1091/mbc.e04-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, et al. A foldable CFTR{Delta}F508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167(6):1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126(3):571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 52.El Khouri E, Le Pavec G, Toledano MB, Delaunay-Moisan A. RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2013;288(43):31177–31191. doi: 10.1074/jbc.M113.470500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, et al. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell. 2008;19(4):1328–1336. doi: 10.1091/mbc.e07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehnert M, Sommermeyer F, Berger M, Kumar Lakshmipathy S, Gauss R, Aebi M, et al. The interplay of Hrd3 and the molecular chaperone system ensures efficient degradation of malfolded secretory proteins. Mol Biol Cell. 2015;26(2):185–194. doi: 10.1091/mbc.E14-07-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16(1):77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 56.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143(4):579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3(1):100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 58.Okiyoneda T, Veit G, Sakai R, Aki M, Fujihara T, Higashi M, et al. Chaperone-independent peripheral quality control of CFTR by RFFL E3 ligase. Dev Cell. 2018;44(6):694–708. doi: 10.1016/j.devcel.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng J, Guggino W. Ubiquitination and degradation of CFTR by the E3 ubiquitin ligase MARCH2 through its association with adaptor proteins CAL and STX6. PLoS ONE. 2013;8(6):e68001. doi: 10.1371/journal.pone.0068001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen HY, Liu CC, Chen RH. Cul3-KLHL20 ubiquitin ligase: physiological functions, stress responses, and disease implications. Cell Div. 2016;11:5. doi: 10.1186/s13008-016-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin DH, Yue P, Yarborough O, 3rd, Scholl UI, Giebisch G, Lifton RP, et al. Src-family protein tyrosine kinase phosphorylates WNK4 and modulates its inhibitory effect on KCNJ1 (ROMK) Proc Natl Acad Sci USA. 2015;112(14):4495–4500. doi: 10.1073/pnas.1503437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE. 2008;3(1):e1487. [DOI] [PMC free article] [PubMed]
- 63.Veit G, Oliver K, Apaja PM, Perdomo D, Bidaud-Meynard A, Lin ST, et al. Ribosomal stalk protein silencing partially corrects the DeltaF508-CFTR functional expression defect. PLoS Biol. 2016;14(5):e1002462. doi: 10.1371/journal.pbio.1002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ameen NA, Marino C, Salas PJ. cAMP-dependent exocytosis and vesicle traffic regulate CFTR and fluid transport in rat jejunum in vivo. Am J Physiol Cell Physiol. 2003;284(2):C429–C438. doi: 10.1152/ajpcell.00261.2002. [DOI] [PubMed] [Google Scholar]
- 65.Ameen N, Silvis M, Bradbury NA. Endocytic trafficking of CFTR in health and disease. J Cyst Fibros. 2007;6(1):1–14. doi: 10.1016/j.jcf.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prince LS, Workman RB, Jr, Marchase RB. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci USA. 1994;91(11):5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noh SH, Gee HY, Kim Y, Piao H, Kim J, Kang CM, et al. Specific autophagy and ESCRT components participate in the unconventional secretion of CFTR. Autophagy. 2018;14(10):1761–1778. doi: 10.1080/15548627.2018.1489479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bodas M, Vij N. Adapting proteostasis and autophagy for controlling the pathogenesis of cystic fibrosis lung disease. Front Pharmacol. 2019;10:20. doi: 10.3389/fphar.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng JW, Zeng XL, Li FY, Ma MM, Yuan F, Liu J, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) prevents apoptosis induced by hydrogen peroxide in basilar artery smooth muscle cells. Apoptosis. 2014;19(9):1317–1329. doi: 10.1007/s10495-014-1014-z. [DOI] [PubMed] [Google Scholar]
- 70.Taibi A, Mandavawala KP, Noel J, Okoye EV, Milano CR, Martin BL, et al. Zebrafish churchill regulates developmental gene expression and cell migration. Dev Dyn. 2013;242(6):614–621. doi: 10.1002/dvdy.23958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kok FO, Shepherd IT, Sirotkin HI. Churchill and Sip1a repress fibroblast growth factor signaling during zebrafish somitogenesis. Dev Dyn. 2010;239(2):548–558. doi: 10.1002/dvdy.22201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Londin ER, Mentzer L, Gates KP, Sirotkin HI. Expression and regulation of the zinc finger transcription factor Churchill during zebrafish development. Gene Expr Patterns. 2007;7(6):645–650. doi: 10.1016/j.modgep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115(5):603–613. doi: 10.1016/S0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- 74.Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun. 2018;9(1):1366. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011;9(4):e1001046. [DOI] [PMC free article] [PubMed]
- 76.Consortium EP The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 77.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47(W1):W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernardo GM, Bebek G, Ginther CL, Sizemore ST, Lozada KL, Miedler JD, et al. FOXA1 represses the molecular phenotype of basal breast cancer cells. Oncogene. 2013;32(5):554–563. doi: 10.1038/onc.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest. 2009;119(6):1537–1545. doi: 10.1172/JCI38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hodos RA, Strub MD, Ramachandran S, Li L, McCray PB, Jr, Dudley JT. Integrative genomic meta-analysis reveals novel molecular insights into cystic fibrosis and DeltaF508-CFTR rescue. Sci Rep. 2020;10(1):20553. doi: 10.1038/s41598-020-76347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1. Summary of differentially expressed genes across conditions in CFBE cells. Summary of up- and down-regulated differentially expressed genes in conditions vs. controls. Table S2. Differentially expressed genes in the miR-138/SIN3A condition. List of differentially expressed genes in the miR-138/SIN3A conditions vs. scrambled siRNA control. Table S3. Differentially expressed genes in the NEDD8/SYVN1 condition. List of differentially expressed genes in the NEDD8/SYVN1 conditions vs. scrambled siRNA control. Table S4. Differentially expressed genes in the temperature condition. List of differentially expressed genes in the temperature conditions vs. 37°C control. Table S5. CFTR interactome used as seed nodes. List of CFTR effectors and interactors used as seed nodes in the M-module analysis. Table S6. DsiRNA and primer sequences. List of siRNA and primer sequences used in the functional knockdown experiments to test for CFTR rescue. Table S7. Untested non-seed module genes. List of genes resulting from the M-module analysis that have not been previously tested or linked to CFTR. Table S8. Top 50 predicted gene ontology biological processes for CHURC1. List of biological processes associated with CHURC1 according to the ARChS4 software. Table S9. Top 50 predicted gene ontology biological processes for RPL15. List of biological processes associated with RPL15 according to the ARChS4 software. Table S10. Top 50 predicted gene ontology biological processes for GZF1. List of biological processes associated with GZF1 according to the ARChS4 software. Figure S1. Schematic showing intersection of differentially expressed genes across conditions. Controls and conditions are described in Table 1. Up arrows indicate up-regulated genes; down arrows indicate down-regulated genes. Significance is defined as FDR < 0.05. Figure S2. Representative transepithelial current tracings demonstrating the effects of individual gene knockdown on CFTR-dependent chloride current in CFBE cells. The Y-axis represents transepithelial current in µA and the X-axis represents time in seconds. The addition of the cAMP agonists forskolin and IBMX resulted in an increase in CFTR-dependent transepithelial chloride current in cells treated with DsiRNAs targeting: A) CHURC1 or B) RPL15. This increase in current was inhibited by the CFTR channel inhibitor GlyH-101. The tracing shown in C demonstrates that DsiRNA knockdown of THOC7 was ineffective in restoring CFTR-dependent chloride current.
Data Availability Statement
The datasets analyzed during the current study are available in the Gene Expression Omnibus Series GSE142610 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142610) [82]. M-module software can be installed at https://github.com/tanlabcode/M-module. Individual modules and datasets are available from the corresponding author on reasonable request.