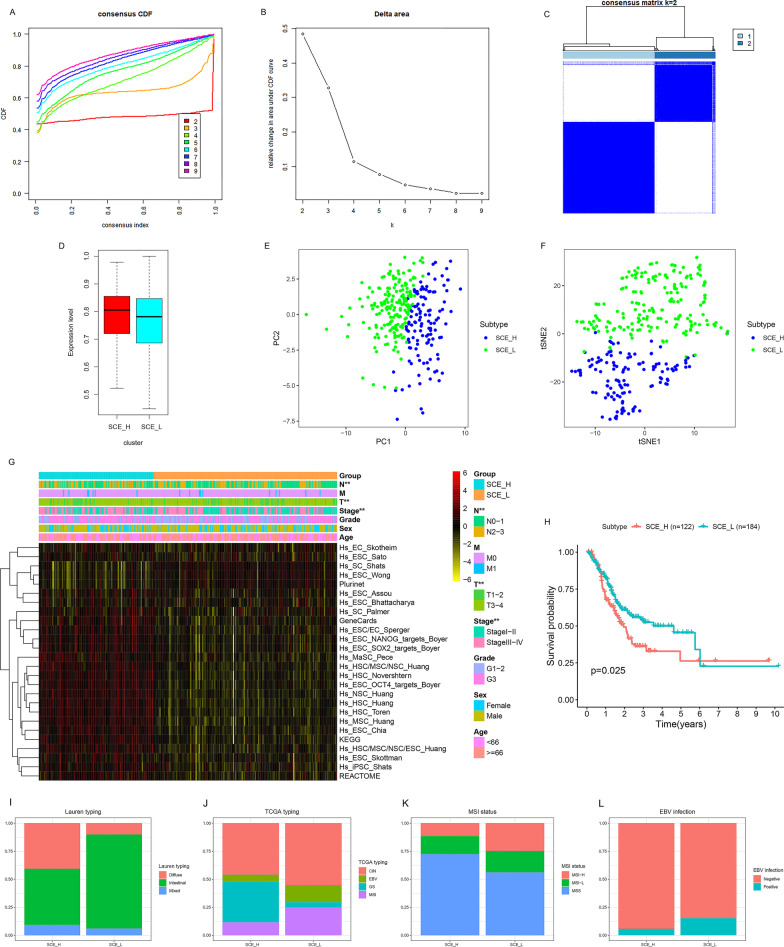
Fig. 1.
Identification of GC stem cell subtypes based on the TCGA database. a–c Two stable stem cell subtypes were identified using consensus clustering analysis according to the K-means algorithm and CDF curve. d GC subtypes were classified as SCE_L and SCE_H based on 26 stem cell gene sets. Clustering of patients belonging to SCE_L and SCE_H in the TCGA cohort based on e PCA and f tSNE algorithm. g The expression of 26 stem cell gene sets and the proportion of clinicopathological features in SCE_L and SCE_H. h Kaplan–Meier analysis of GC stem cell subtypes. i–l The proportion of i Lauren subtypes, j TCGA subtypes, k MSI subtypes, and l EBV infection subtypes in GC stem cell subtypes. Statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001