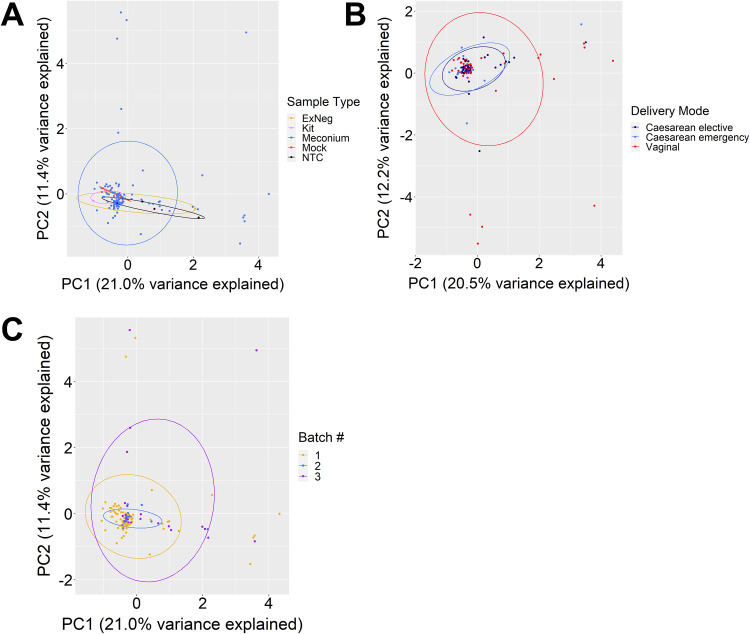
FIG 2.
Microbiome profiles of meconium show no compositional differences compared to sequencing negative controls and are not affected by delivery mode or sequencing batch. Principal-component analysis (PCA) of log ratio transformed cpn60 read count data grouped by sample type, delivery mode, and extraction kit batch number. (A) PERMANOVA reported no significant separation by sample type across the entire data set (P = 0.923, r2 = 0.02) and no significant differences in between-group dispersion (P = 0.246, F = 1.33). (B) Meconium samples were reported to cluster significantly by delivery mode (P < 0.05, r2 = 0.03); however, permutation of dispersion data also showed a significant difference in between-group dispersion (P < 0.05, F = 4.76). (C) Samples across the entire data set also clustered significantly by sequencing run (P < 0.001, r2 = 0.03) with significant differences in dispersion between groups (P < 0.01, F = 4.23). Lack of evident separation of samples by delivery mode and extraction kit batch number on the respective plots, combined with permutation analysis of dispersion data, indicates that the “significant” clustering results should be interpreted as statistical artifacts rather than true biologically important differences.