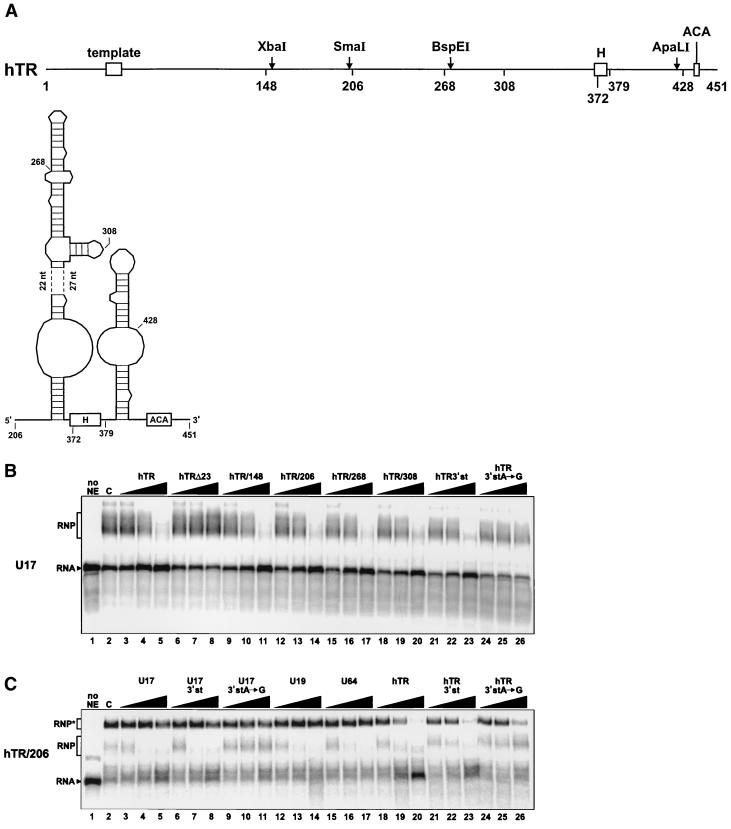
FIG. 5.
hTR and its derived 3′-terminal fragments can compete for U17 assembly. (A) Schematic structure of hTR RNA (nt 1 to 451) and secondary structure model of its H/ACA-like domain. Open boxes represent the template region and the H and ACA motifs. Relevant restriction sites are indicated by an arrow. Numbers indicate the first nucleotides of 5′-truncated hTR RNA fragments. The secondary structure model was adapted from reference 47. (B) Gel mobility shift assay in the presence of competitor RNAs hTR and its derived RNA fragments. Radiolabeled U17 RNA was incubated in the absence (lane 1) or presence (lane 2) of WCE. Increasing amounts (10-, 100-, and 1,000-fold molar excess) of unlabeled competitor RNAs hTR (lanes 3 to 5), hTRΔ23 (lanes 6 to 8), hTR/148 (lanes 9 to 11), hTR/206 (lanes 12 to 14), hTR/268 (lanes 15 to 17), hTR/308 (lanes 18 to 20), hTR/3′st (nt 378 to 451; lanes 21 to 23), and hTR/3′stA→G (lanes 24 to 26), which contains an A-to-G mutation in the ACA motif, were added to the assays. (C) Gel mobility shift assay with radiolabeled hTR/206. The labeled RNA was incubated in the absence (lane 1) or presence (lane 2) of WCE and could form two RNP complexes, designated RNP and RNP∗ for the faster- and the slower-migrating complexes, respectively. Unlabeled competitor RNAs U17 (lanes 3 to 5), U17/3′st (lanes 6 to 8), U17/3′stA→G (lanes 9 to 11), U19 (lanes 12 to 14), U64 (lanes 15 to 17), hTR (lanes 18 to 20), hTR/3′st (lanes 21 to 23) and hTR/3′stA→G (lanes 24 to 26) were added in increasing concentrations (10-, 100-, and 1,000-fold molar excess).