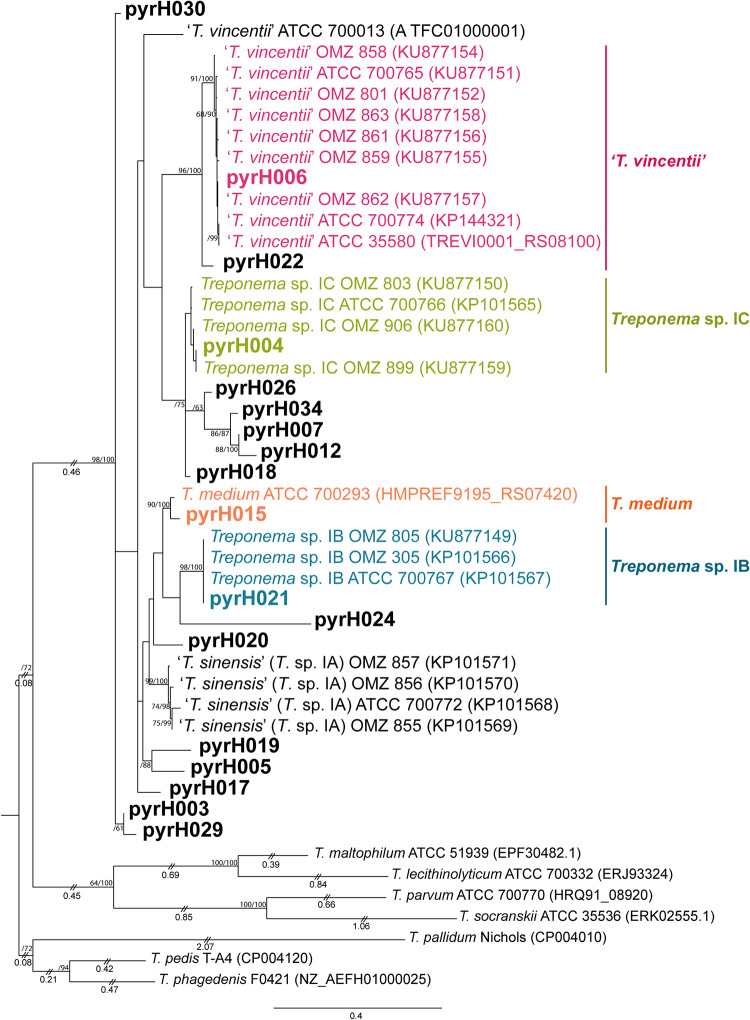
FIG 3.
Maximum-likelihood (ML) phylogenetic tree of pyrH genes from oral treponeme phylogroup 1 taxa. The maximum clade credibility tree topology is supported by bootstrapping for 1,000 replicates (first number) and Bayesian posterior probabilities (second number) reported as percentage values separated with a forward slash (“/”) beside the branch nodes. Only values over 60% are shown. Treponeme genotypes identified in this study are indicated in boldface. Excessively long branches have been trimmed in proportion to the scale bar (indicated with double slashes [“//”] on the respective branches). The scale bar indicates 0.4 nucleotide changes per position. The oral treponeme species (phylogroups) are indicated with different colors as follows: T. vincentii, pink; T. medium, orange; Treponema sp. IC, yellow-green, and Treponema sp. IB, cyan. Treponema pedis T-A4, Treponema phagedenis F0421, Treponema pallidum Nichols, Treponema socranskii subsp. socranskii ATCC 35536, Treponema parvum ATCC 700770, Treponema maltophilum ATCC 51939, and Treponema lecithinolyticum ATCC 700332 were included as outgroup species.