Abstract
There is compelling evidence that sex and gender have crucial roles in excessive alcohol (ethanol) consumption. Here we review some of the data from the perspective of brain transcriptional differences between males and females, focusing on rodent animal models. A key emerging transcriptional feature is the role of neuroimmune processes. Microglia are the resident neuroimmune cells in the brain, and exhibit substantial functional differences between males and females. Selective breeding for binge ethanol consumption, as well as the impacts of chronic ethanol consumption and withdrawal from chronic ethanol exposure, all demonstrate sex-dependent neuroimmune signatures. A focus is on resolving sex-dependent differences in transcriptional responses to ethanol at the neurocircuitry level. Sex-dependent transcriptional differences are found in the extended amygdala and the nucleus accumbens. Telescoping of ethanol consumption is found in some, but not all, studies to be more prevalent in females. Recent transcriptional studies suggest that some sex differences may be due to female-dependent remodeling of the primary cilium. An interesting theme appears to be developing: at least from the animal model perspective, even when males and females are phenotypically similar, they differ significantly at the level of the transcriptome.
Keywords: binge, ethanol, functional genomics, neural circuitry, neuroimmune function, therapeutics
INTRODUCTION
There is ample evidence of interactions between alcohol use disorder (AUD) and sex (biological factors) and gender (psychological, social, and cultural factors) (1,2). In this review article, we focus on how the interaction between sex and the transcriptome aligns in animal models of excessive ethanol consumption. Wilhelm et al. (3) were among the first to provide compelling evidence for sex-specific effects of alcohol on the brain transcriptome of mice. They found a pro-inflammatory transcriptional phenotype in females during peak withdrawal from chronic alcohol intoxication, while there was a suppression of neuroimmune signaling in males. Since Wilhelm et al. (3), there are now numerous studies, some described below, that have directly or indirectly provided evidence for sex interactions with the brain transcriptome and associated alcohol phenotypes, and are beginning to be defined on neurocircuitry and molecular levels. We begin with brief discussions of sex differences in AUD and the evidence that responses to alcohol are mediated by different circuits in males and females.
SEX DIFFERENCES IN ALCOHOL USE DISORDER
AUD is a chronic relapsing brain disease characterized by compulsive alcohol use, loss of control over alcohol intake, and a negative emotional state when not using alcohol. Risk factors include additive genetic effects (4), age (5), sex (6,7), and numerous environmental influences, especially those affecting mental health status (8,9). For decades, reports have indicated that males consume more alcohol and consume it more often than females. However, recent surveys in the US indicate that female patterns of drinking are changing (10). While men consistently report higher levels of drinking to intoxication and the prevalence of AUD is higher in men, women are drinking more than previously reported (10), are at greater risk for negative consequences of alcohol use (11) and tend to develop AUD in a shorter timeframe (1,12).
A neuroimmune hypothesis for alcohol use progressing to AUD has been proposed (13) and is supported [see (14,15)]. Compared to men, women are particularly sensitive to alcohol-mediated immune impairment, which may be linked to an increased risk for hypertension and damage to the brain, heart, kidney and liver (16–20). AUD is a prototypical complex trait with a strong heritability of ~0.5 (21); however, few studies report quantitative sex differences. Whether the genetic components between sexes overlap is another question. Twin studies report genetic correlations of 0.24 for opposite sex dizygotic twins, rather than the expected 0.5 shared heritability (22). Similarly, risk is higher if a same sex, rather than an opposite sex, relative has an AUD (23).
As with any psychiatric disorder, the question arises as to how to model AUD in animals. Modeling human disease remains controversial (24,25) (2 references) and is further complicated when one considers the sex x model interaction. Crabbe et al. (2013) considered this issue from the perspective of genetic animal models and concluded that, with regard to AUD, the consilience of the human genetic and animal model genetic data is best for alcohol withdrawal and tolerance, major features of an AUD diagnosis. However, just as human studies have failed to address the role of sex in the genetics of AUD, sex x transcriptome and sex x circuitry effects have only recently become a focus of animal model research and are central topics here.
ETHANOL-RELATED BEHAVIORAL AND NEURAL CIRCUIT SEX DIFFERENCES
A reasonably well-replicated observation is that female rodents voluntarily drink more alcohol (ethanol) than males in operant and two-bottle choice tests (26–31). However, at least in some rodent populations, further increases in consumption after chronic ethanol access are less consistently observed in females than they are in males (32,33). Relative to males, female rodents exhibit reduced withdrawal signs during abstinence after ethanol exposure (33,34), and enhanced sensitivity to rewarding effects (e.g., ethanol-conditioned place preference) (35), but blunted sensitivity to aversive effects (e.g., ethanol-conditioned taste aversion) of ethanol (36). This constellation of behavioral differences – higher sensitivity to the positive and lower sensitivity to the negative effects of ethanol – predicts that female rodents would be at heightened susceptibility to excessive ethanol intake.
The extent to which these sex differences relate to divergent brain anatomy and neurocircuitry function remain unanswered questions. For example, unlike humans, mice have no appreciable sex difference in overall brain weight; males: 458.2 ± 41.6 mg, N=10226; females: 459.7± 42.0 mg, N=10704 (37). In terms of specific brain regions, in mice, there again appear to be minimal sex differences in volumes, with the exception of sub-regions in and around the hypothalamus that are directly related to the modulation of sexual and reproductive behaviors (38–40). Relevant to discussions in subsequent sections is the role of neuroimmune processes in the anatomical and functional divergence in the hypothalamus and the functional divergence in other brain areas (38,39).
Some recent studies are beginning to provide functional context to differences between the sexes in ethanol responses (41). For example, chemogenetic excitation of the nucleus accumbens (NAc) reduces binge drinking in female mice (42,43), whereas chemogenetic inhibition of this region reduces binge drinking in male mice (44); (Townsley, Borroego, and Ozburn, unpublished observations). Of further note, sex-dependent effects of ethanol on neuronal activity have been observed in the basolateral and central nuclei of the amygdala (45–47). Because the accumbens and amygdala are known neural hubs for rewarding and aversive behaviors, they are a plausible focus for sex differences in ethanol behaviors. Further studies are needed to determine how ethanol-related adaptations in the NAc and amygdala may relate to sex-dependent changes, including transcriptional changes in other limbic brain regions, as well as the reciprocally connected prefrontal cortex (PFC) (48).
THE TRANSCRIPTOME, ETHANOL RESPONSES AND THE ESTRUS CYCLE
Prendergast et al. (49), after reviewing 294 studies that included both male and female rodents, concluded that the main reasons females were not included in biomedical research “…is based on the assumption that females are intrinsically more variable than males and must be tested at all stages of the estrus cycle to generate reliable data. Neither belief is empirically based” (emphasis added). From the perspective of this review, we need to ask if the marked differences repeatedly found between males and females in ethanol responses are always much greater than the differences that could be solely associated with estrus cycle effects. The behavioral data are not clear on this point. Forger and Morin (50) reported that in Sprague-Dawley rats, ethanol intake and preference over the 4 day estrus cycle varied systematically, being lowest on the day of proestrus, a decrease of > 50% from the peak value. In contrast, Priddy et al. (51) found significant differences between male and female Wistar and Long-Evans rats in voluntary ethanol consumption (two-bottle choice), but no significant influence of the stage of the estrus cycle on total ethanol consumption. To add to the complexity, Ford et al. (52) dissected ethanol consumption into discrete bouts and reported that the patterns of ethanol consumption by female Long-Evans rats differ over the course of the estrus cycle. Ford et al. (52) concluded that, “the estrus cycle phase-related changes in microstructural components of ethanol intake suggest that female rats experience periods of altered sensitivity to the neurobiological and reinforcing effects of ethanol.”
There are to our knowledge no studies trying to align changes in the transcriptome and ethanol-associated behaviors with the estrus cycle. In fact, studies looking at brain gene expression across the estrus cycle are relatively rare. DiCarlo et al. (53) found that across four brain regions (hippocampus, neocortex, hypothalamus and cerebellum) in C57BL/6J (B6) mice (a high ethanol preference strain) there were only 210 differentially expressed genes out of 16,000 genes detected as present; 61 of these genes were known to be estrogen responsive. These genes were enriched in annotations of myelination, hormone stimulus and abnormal hormone levels. Duclot and Kabbaj (54) examined gene expression in the medial PFC of young (8 week) male and female Sprague-Dawley rats; importantly, the female data were collected across the estrous cycle. There were 935 genes differentially expressed from proestrus to diestrus and many of these genes were associated with the regulation of synaptic activity (52). The differences between these two studies may reflect species differences, differences in the brain regions surveyed and/or technical differences.
To further probe the issue of brain gene expression across the estrus cycle, RNA-Seq was used to examine the effects of the estrus cycle in two rat strains (BN-Lx/Cub and SHR/OlaIpcv). Data were collected in pre-estrus, estrus and post-estrus (experimental details are found in Supplemental Information Tables S1 and S2). The number of transcripts for which expression levels were affected by stage of the estrus cycle ranged from 4 to 262, depending on p-value threshold (0.0001 to 0.05). Because of the short period of each stage of the estrus cycle in the rat, the technique that we used to determine the stage of the cycle should be supplemented with values of circulating hormones in the future. The differences that we noted at various levels of significance should stimulate research in this area using more definitive tools to denote the hormonal state of the animal at the time that tissue is obtained. Additionally, investigation of individual splice variants, alternative polyadenylation events, and levels of RNA expression in particular cell types will provide important information relating estrus stage to transcriptional differences.
TRANSCRIPTOMICS REVEALS SEX X STRAIN DIFFERENCES IN THE RESPONSE TO ACUTE ETHANOL INTOXICATION.
For over six decades, a staple of ethanol research has been rodent inbred strains that differ in their voluntary consumption of and responses to ethanol. For example, DBA/2J (D2) mice drink little ethanol and exhibit severe ethanol withdrawal symptoms, whereas B6 mice drink excessive amounts and exhibit mild withdrawal (55). The genetic relationship of low withdrawal symptomatology with high ethanol consumption has been demonstrated (55). In the standard two-bottle choice test, B6 females consistently consume more ethanol (g/kg) than males. These strain differences were leveraged in the first meta-analysis of brain gene expression data for any behavioral phenotype (56). The transcriptome of high and low ethanol preference animals differed markedly, and mitogen-activated protein kinase signaling, transcription regulation pathways and cytokine signaling pathways were significantly different. Female expression data were included, but the data were not analyzed for sex differences. Over the past 15 years, females have increasingly been included in ethanol research due to the recognition that sex differences in brain gene expression sometimes exceed strain differences and/or the data support significant sex x genotype interaction. An example of this point is provided here, for the effects on whole brain gene expression 7 h after a 4 g/kg ethanol dose vs. saline in female and male B6 and D2 mice [N=10/group, see Supplemental Information Table S3 for expression data; analysis details are similar to our previous publication (57)].
Given that differences between strains and/or sexes may be due to significant differences in variance structure, the variances for 9911 genes meeting the threshold for analysis inclusion were calculated. Two-way AVOVAs (sex x treatment) for variance data were completed for each strain. No significant differences in variance were found. Venn diagrams (Figure 1) illustrate that between 10–15% of total gene expression was changed by ethanol treatment (p < 0.05), with the greatest change seen in D2 males. The overlap in differential gene expression between the sexes was minimal at approximately 10%. Principal components analyses found divergent sex × treatment components for both the B6 and D2 strains (Figure 2). KEGG results from WebGestalt (58) analyses indicated several strain distinct pathways with significant sex × treatment divergence (Table 1). Consistent sex differences in ethanol-modulated pathways, regardless of strain, were detected by Ingenuity Pathway Analysis (IPA) and are shown in (Table 2). The same gene input was used for both, with IPA also including fold-change data.
Figure 1.
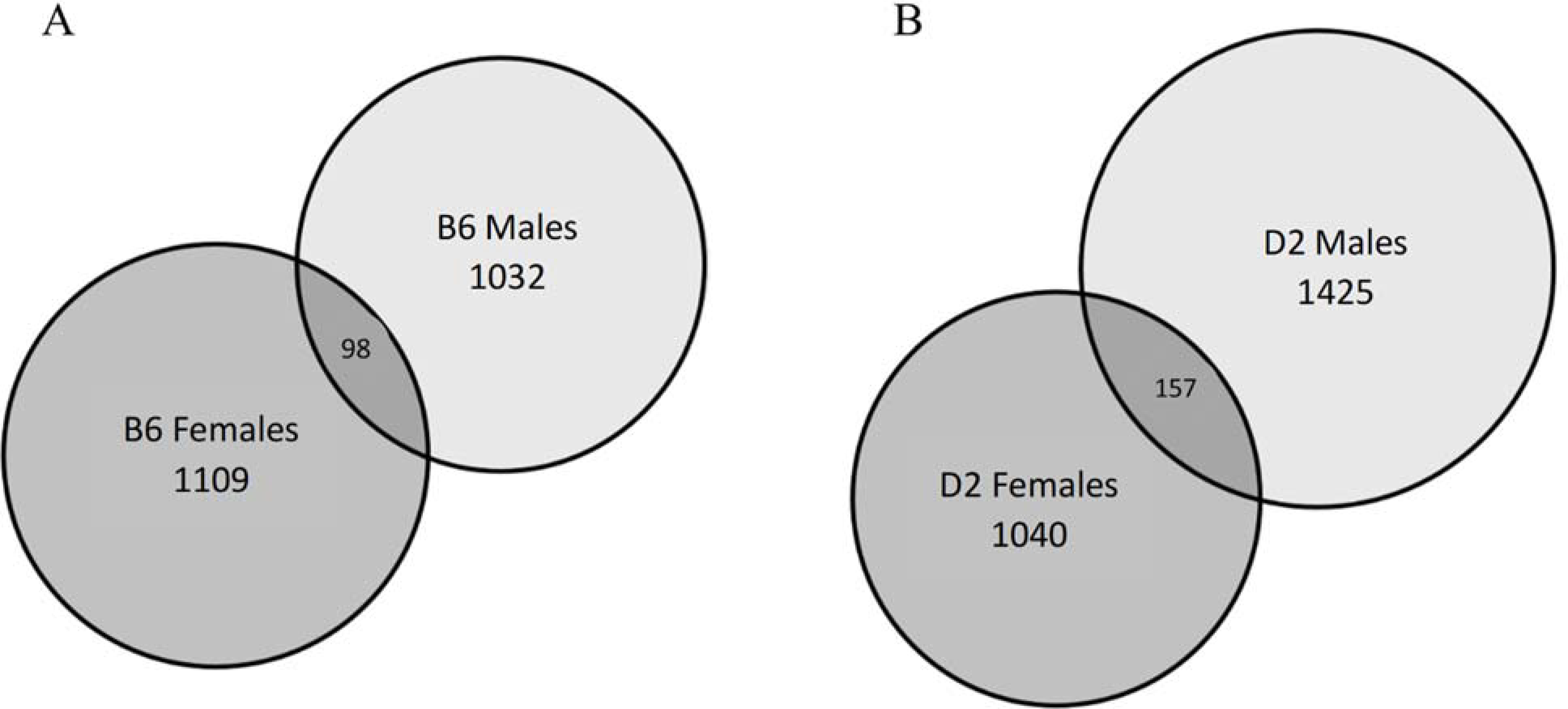
Minimal overlap of ethanol-mediated gene expression changes between sexes are represented by Venn diagram. Numbers within the circles represent significant (p<0.05) ethanol-mediated expression changes. A) C57BL/6J (B6) and B) DBA/2J (D2) mice. Both strains show similar patterns, with a higher number of changes in males than females and low overlap. Left circle: female-specific expression; right circle: male-specific expression; overlapping dark gray: ethanol-mediated changes in both sexes. n=10/per sex, strain and treatment; 80 total cDNA arrays
Figure 2.
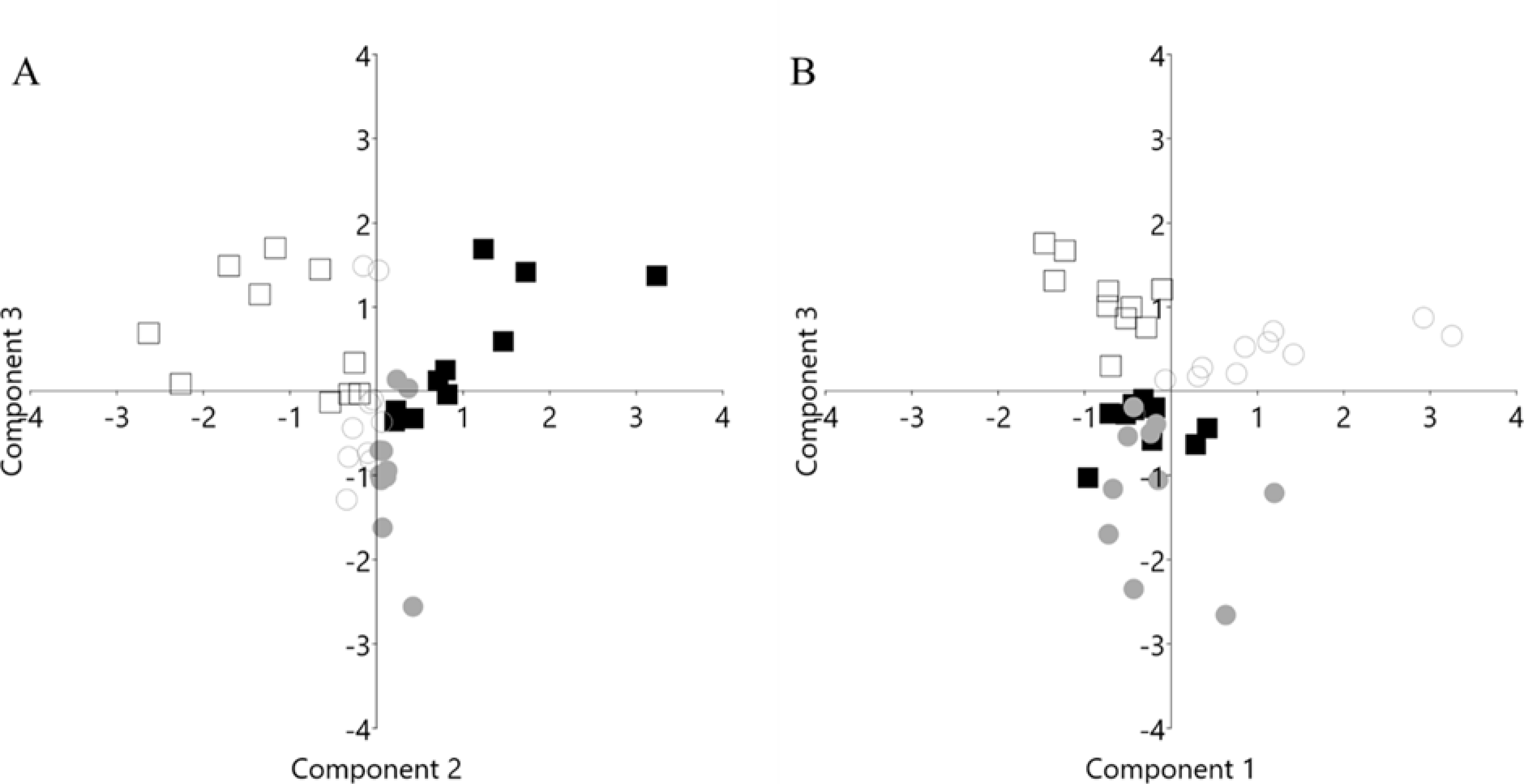
Divergence of gene expression by sex and ethanol within strain is shown by principal components analyses (PCA). PCA for A) C57BL/6J and B) DBA/2J gene expression are shown in eigenvalue scale; 9911 genes were entered into the analyses. Circles: female, squares: male; filled: control; open: ethanol. n=10/per sex, strain and treatment; 80 total cDNA arrays
Table 1.
KEGG analysis. Sex differences in over-represented pathways by mouse strain.
| Female | Male | |||
| C57BL/6J Pathway Analysis | p-value | FDR | p-value | FDR |
|
| ||||
| Ubiquitin mediated proteolysis | 7.67E-06 | 0.002 | 1.000 | 1.000 |
| Relaxin signaling pathway | 1.000 | 1.000 | 7.4E-06 | 0.002 |
| Pathways in cancer | 0.106 | 0.524 | 0.000168 | 0.027 |
| Cholinergic synapse | 0.680 | 0.980 | 0.000395 | 0.042 |
| PPAR signaling pathway | 0.760 | 1.000 | 0.000649 | 0.049 |
| Fc gamma R-mediated phagocytosis | 1.000 | 1.000 | 0.000814 | 0.049 |
| Colorectal cancer | 1.000 | 1.000 | 0.000908 | 0.048 |
| DBA/2J Pathway Analysis | p-value | FDR | p-value | FDR |
|
| ||||
| Glycosphingolipid biosynthesis | 5.81E-05 | 0.012 | 0.126 | 0.519 |
| Focal adhesion | 7.34E-05 | 0.012 | 0.009 | 0.133 |
| Insulin resistance | 0.000121 | 0.013 | 1.000 | 1.000 |
| TGF-beta signaling pathway | 0.000306 | 0.020 | 1.000 | 1.000 |
| FoxO signaling pathway | 0.000378 | 0.020 | 0.006 | 0.131 |
| Autophagy | 0.000923 | 0.039 | 0.085 | 0.386 |
| Adherens junction | 0.000981 | 0.039 | 1.000 | 1.000 |
| Colorectal cancer | 0.035 | 0.198 | 1.21E-05 | 0.002 |
| Bacterial invasion of epithelial cells | 0.034 | 0.197 | 1.46E-05 | 0.002 |
| Endocytosis | 1.000 | 1.000 | 2.07E-05 | 0.002 |
| Protein processing in endoplasmic reticulum | 1.000 | 1.000 | 3.63E-05 | 0.003 |
| Spliceosome | 0.018 | 0.143 | 0.000511 | 0.033 |
| Pancreatic cancer | 0.014 | 0.128 | 0.000815 | 0.040 |
| ErbB signaling pathway | 0.137 | 0.444 | 0.000872 | 0.040 |
Significant expression was determined from cDNA arrays; p<0.05, n=10/per sex, strain (C57BL/6J and DBA2J) and treatment (control vs binge ethanol); 80 total cDNA arrays. The input was gene symbols.
Table 2.
IPA analysis. Consistent sex differences in over-represented pathways regardless of mouse strain.
| p-value | ||||
|---|---|---|---|---|
| IPA Pathway | D2 F | D2 M | B6 F | B6 M |
|
| ||||
| Adipogenesis | 0.000 | - | 0.007 | - |
| Cell cycle: G2/M DNA damage checkpoint regulation | 0.003 | - | 0.010 | - |
| Amyloid processing | 0.011 | - | 0.036 | - |
| Super pathway of inositol phosphate compounds | 0.038 | - | 0.010 | - |
| D-myo-inositol-5-phosphate metabolism | 0.041 | - | 0.005 | - |
| Semaphorin neuronal repulsive signaling | - | 0.038 | - | 0.001 |
| RhoA signaling | - | 0.000 | - | 0.004 |
The input was gene symbols and fold-change using the same data from Figure 1. B6, C57BL/6J; D2, DBA/2J; F, female; M, male
The data summarized above illustrate that the hypnotic dose of ethanol affected up to 15% of genes expressed in each comparison, a level of difference commonly seen in rodent ethanol-related studies (56) However, there was little overlap in female- and male-specific differential gene expression, nor in the over-represented pathways to which they belong. These data affirm the marked differences in transcriptional responses to ethanol between males and females.
SEX, SELECTION, CHRONIC ETHANOL CONSUMPTION AND THE TRANSCRIPTOME
Iancu et al. (59) compared the ventral striatal (the ventral striatum includes the NAc) transcriptome of ethanol naïve High Drinking in the Dark (DID) selected line mice (60) with the genetically heterogeneous stock, HS/NPT founders. The HDID mice drink to intoxicating blood ethanol concentrations (BECs) of > 150 mg of ethanol/dL. Two key parameters extracted were differential expression and differential variability. The number of significant (FDR < 0.05) differentially expressed genes in females and males was 227 and 1525, respectively; 153 genes overlapped. No significant gene ontology (GO) annotation was detected for the genes affected in females. For males, 836 and 689 genes were down- and up-regulated, respectively. Annotation of the down-regulated genes revealed significant enrichment in genes associated with extracellular matrix organization, immune system process, collagen trimer and plasma membrane part. For the up-regulated genes, no significant enrichment categories were detected.
For the differential variability metric, selection significantly (FDR < 0.05) increased the variability of 1498 genes for females and 766 genes for males; 82 genes overlapped. Included in the overlapping subset were Calb2, Gabrq, Nos1ap, Oxt, Pomc, Pvab, Slc6a11 and Trh. For genes with increased variance in females (N=1418), there was significant enrichment in annotations that included extracellular space, plasma membrane part, signaling receptor activity, and extracellular matrix organization. For genes with decreased variance in females (N=80), significant enrichment was detected for cytoskeleton of presynaptic active zone and axon part; genes involved included Bsn, Pclo, Syn1, Myoc, Nav1, Tubb4a, Cplx2 and Ank3. For genes with increased variance in males (N=663), there were significant enrichments in GO categories that included modulation of synaptic transmission, voltage gated cation channel activity, plasma membrane part, and synapse part. Genes in the latter category included Grin2a, Grin2b, Dlg4, Gabbr2, Grm2, Pdyn, Gabra1 and Camk2a. For genes with decreased variance in males (N=103), there were significant enrichments in GO categories associated with biological adhesion and extracellular part. From the perspective of the differential variability metric, which is closely positively aligned with intra-modular connectivity (61), the female and male data are largely mirror images.
Hitzemann et al. (62) examined the effects of chronic (13 weeks) ethanol consumption [using a two-bottle choice (water vs 10% ethanol) design] on CeA gene expression in heterogeneous stock-collaborative cross (HS-CC) mice. The CeA is a key region for sustaining high ethanol preference (63). The critical parameter discussed here is the correlation of individual gene expression and ethanol preference (during final drinking week 13). For females, the genes significantly positively associated with preference (N=536) were enriched in annotations associated with cilium movement, cilium organization, extracellular region, and collagen-containing extracellular matrix. There were no significant annotations in females for the negatively correlated genes, nor in males, for the positively or negatively correlated genes.
The weighted gene correlation network analysis (WGCNA) (64) was used to form a female gene expression network. The majority of genes that correlated with preference (N=376) were found in a single network module. This module was enriched (p < 0.0001) in genes with an astrocyte annotation [see (65)]. Annotation included extracellular matrix and cilium. Among the positively correlated genes, 43 had a relative intra-modular connectivity ≥ 0.9; i.e., they were top hub nodes. Enrichr (66,67) was used to search for key transcription factors among the top hub nodes. A key finding was that 19 of the top nodes were down-regulated in an orthodenticle homeobox 2 (Otx2) knockout mouse (GSE27630; (68)). Otx2 was one of the positively correlated top hub nodes from the WGCNA for the female gene expression network, and often is referred to as a master regulator, known to have key roles in brain patterning and postnatal plasticity. Otx2 is further required for generation of various neuronal subpopulations, including ocular motor and midbrain dopaminergic neurons (68,69), and development and maintenance of perineuronal nets. In the adult brain, Otx2 expression is largely localized to the choroid plexus (70) and the OTX2 protein is captured by the perineuronal nets and accumulated in parvalbumin type GABA-ergic neurons throughout the brain (71). Our data indicate a low, but detectable expression of Otx2 in the CeA, affected by ethanol exposure and predicted to have a role in the escalation of ethanol preference seen in HS-CC females, but not males, and in the observed sex differences in the transcriptional response.
SEX DIFFERENCES IN THE INNATE NEUROIMMUNE TRANSCRIPTOME AND CHRONIC ETHANOL CONSUMPTION.
The brain innate immune system recognizes known molecular patterns to produce a rapid inflammatory response in microglia. Importantly for this review, microglia show marked functional sex differences (see e.g., (72)). The neuroimmune hypothesis of AUD, predicts that alcohol consumption promotes innate pro-inflammatory signaling, which leads to increased alcohol intake, creating a positive feedback loop (73). Recent evidence suggests that sex differences in this process are localized at the level of the microglia (74). The exact mechanisms associated with the sex-dependent effects are unclear.
The innate immune system can be activated experimentally by administration of small molecule ligands of Toll-like receptors (TLRs) such as Poly(I:C) (PIC) or lipopolysaccharide (LPS). Several TLR agonists have been shown to modulate ethanol consumption (75–78), but the effects are complex. LPS caused a reproducible increase in chronic ethanol intake in male and female B6 mice, though a continued increase in ethanol consumption was found one month after LPS treatment in males, but not females (75). When female FVB, FVBxB6F1 and B6xNZBF1 mice were tested, the F1, but not FVB mice exhibited the LPS-induced escalation of drinking (75). Warden and colleagues (76,77) administered repeated every other day PIC injections and found a robust increase in ethanol intake in B6 males, but a similarly robust decrease in B6 females. When the frequency or the time point of PIC injections was changed, males decreased their ethanol consumption, while a decrease was no longer found for females, suggesting a strong influence of sex on the time-course of immune responses and behavior. Time-dependent analyses demonstrated that the peak immune activation in B6 males occurred earlier than in B6 females and the authors hypothesized that availability of ethanol during peak activation would result in decreased drinking, whereas ethanol consumption during the descending limb of immune activation may increase consumption.
Using Camk2a-Sun1F1 hybrid male and female mice (see (79) for genotype detail) and a modification of a previously published procedure (76), repeated injections of PIC did not change ethanol consumption in females, but there was a trend for reduced drinking in males (p < 0.1, data not shown). Immune gene expression profiles for the frontal cortex (Figure 3) indicated that innate immune activation by PIC resulted in a robust increase in expression of pro-inflammatory cytokines 3 hours after the last PIC injection with a smaller increase or a return to baseline at the 48 h time point. Further, F1 males and females were largely similar in their immune response. These data, and those of Warden et al. (76,77,80), illustrate that the immune modulation of ethanol consumption in mice is strongly influenced by sex and genotype and that the sex- and genotype-specific effects depend on the time-course of neuroimmune activation.
Figure 3.
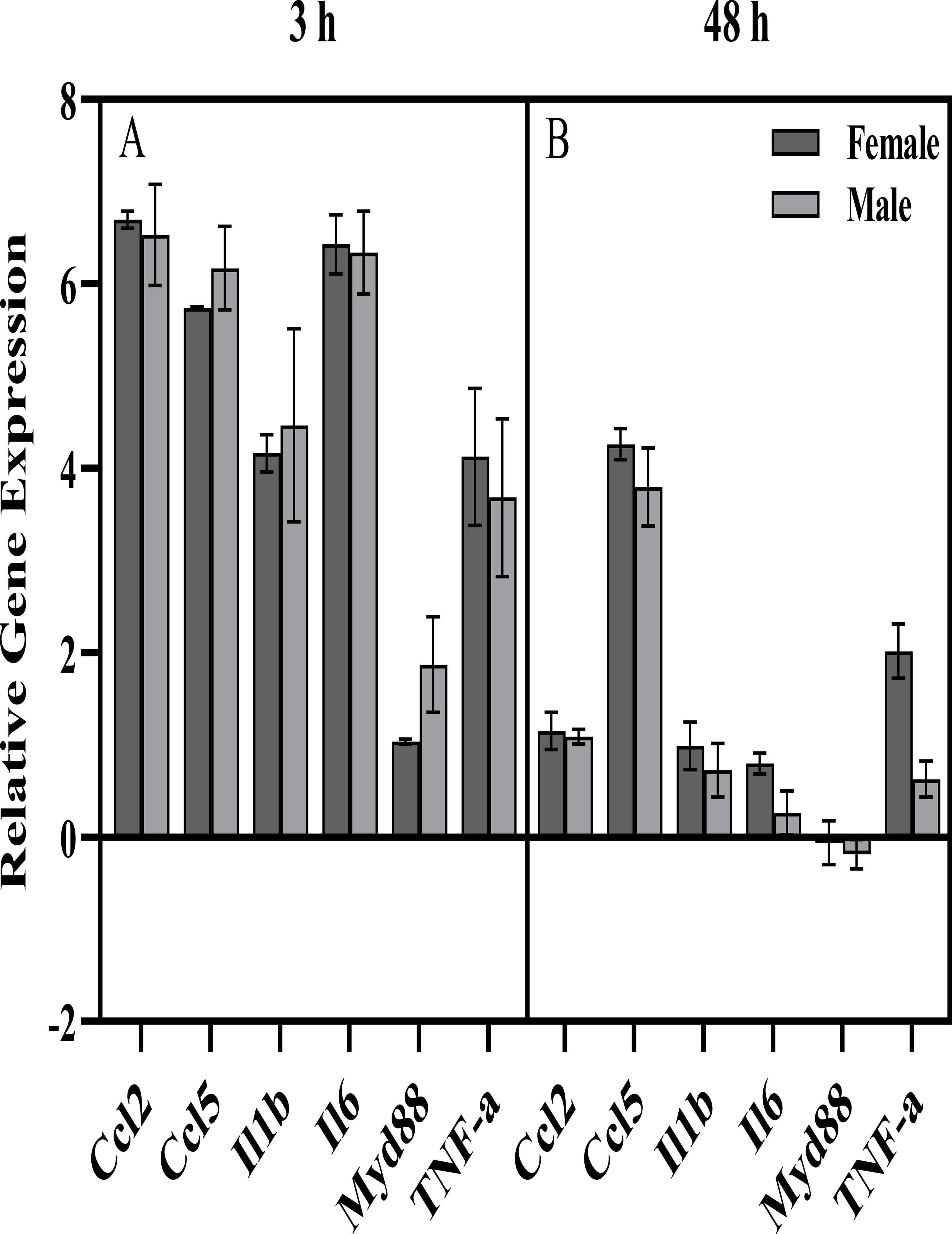
Expression of immune genes in frontal cortex of Camk2a-Sun1 F1 mice A) 3 h and B) 48 h after an injection of Poly(I:C) [PIC] following every other day alcohol consumption. These mice are created by crossing the Camk2a-Cre (https://www.jax.org/strain/005359) and CAG-LSL-Sun1-sfGFP-Myc (https://www.jax.org/strain/021039) mouse lines, which have been used to study cell type-specific responses to perturbations (79). A modification of a previously published behavioral procedure was used (76). Briefly, male and female mice (n=4–7 per group, 8–11 weeks old at the start of the experiment) were injected i.p. with either saline or PIC (10 mg/kg) every 4 days for a total of 13 injections. Ethanol (15% v/v) was available every other day in a two-bottle choice procedure (with water), starting 24 hours after the first PIC injection. Mice were euthanized and frontal cortex was dissected for molecular analysis 3 h or 48 h after the last PIC injection. Expression of 6 genes including 4 pro-inflammatory cytokines, Il6 and Myd88 was measured using qRT-PCR (TaqMan®). Gene Expression was normalized to the saline control of each sex and geometric mean of GusB and 18s (-ddCt, Log2 scale). Error bars represent SEM. Sex x Time ANOVAs revealed significant effects of Time for all genes, with generally lower expression at the 48 h time point, compared to 3 h, but no significant effects of Sex or Sex x Time interactions. One-tailed t-tests comparing PIC group to saline baseline found a significant increase for all genes (except Myd88) of both sexes at the 3 h time point, as well as a significant increase for 4 out of 6 genes for females (Ccl2, Ccl5, Il1b, Il6, TNF-a), and a significant increase for 1 out of 6 genes for males (Ccl5) at the 48 h time point (all p<0.05 Bonferroni-corrected for multiple comparisons).
SEX DIFFERENCES IN THE NEUROIMMUNE TRANSCRIPTOME AND BINGE ETHANOL CONSUMPTION
Binge level alcohol drinking is a hallmark of AUD and can be obtained in mice under intermittent schedules of ethanol access. Recent research characterized gene expression differences in the NAc of adult male and female B6 mice at 24 h after 7 intermittent, binge-like ethanol drinking sessions and compared the results to controls drinking water (81). Average ethanol intake was high (2.34 g/kg/30 min in females; 2.35 g/kg/30 min in males) and BECs (>1.3 mg/mL) exceeded the NIAAA criteria for binge drinking (0.8 mg/mL). Customized qPCR Mouse Mood Disorder arrays quantified expression levels of 384 genes and found marked sex differences for the top 30 genes regulated by binge-like ethanol drinking. Expression differences characterized by IPA suggested that binge level drinking altered hormone signaling and immune function in females and neurotransmitter metabolism in males (81). One canonical pathway of interest was “TNFR2 signaling,” with an overall pattern of gene expression changes consistent with inactivation of the pathway in females and activation of the pathway in males. For example, repeated binge level ethanol drinking produced significant and opposite changes in the expression of Lta. Lta encodes lymphotoxin-α, which is involved in cell survival, proliferation, differentiation, apoptosis, and immune regulation. Repeated binge level drinking also decreased expression of Il6 only in females, and the pattern of the ethanol drinking-induced reduction in some cytokine levels in females is consistent with a decrease in the initiation of signaling at TNFR1 (82) and TNFR2 (81). The fairly consistent down-regulation of signaling through the TNF superfamily following repeated binge-like drinking in females, including the down-regulation of the RelA subunit of NF-ĸB (Rela), likely decreases activation of the transcription factor NF-ĸB. NF-ĸB has also been identified as a central node in male and female PFC gene networks during acute ethanol withdrawal following chronic vapor intoxication (81). However, the interacting gene sets were distinct, with several genes in females indicative of a pro-inflammatory response, and several genes in males related to immunosuppression (3). Consistent with this idea, chronic intoxication activated inflammatory signaling and cell death pathways only in females, and subsequent studies in the PFC confirmed significant neuronal degeneration in females but a reduction in cell death in males (3,83). Thus, the opposite effects of repeated binge level ethanol drinking and chronic intoxication on immune function, with changes in females consistent with a decrease in cell survival and an increase in inflammation and apoptosis, have important implications for a role of neuroimmune signaling in the acute and chronic effects of ethanol, including neurodegeneration (reviewed in (82,84)).
SEX AND TRANSCRIPTOMICS: SEARCHING FOR TARGETS TO MANIPULATE ETHANOL CONSUMPTION
A major initiative is to utilize transcriptome information from ethanol studies to meet treatment and prevention goals. Once genes and gene networks are identified, manipulations can be performed to test hypotheses about their roles in ethanol intake and in genetic risk for higher levels of consumption. For example, ethanol-naïve male HDID-1 and HS/NPT founder mice were used to identify molecular signatures (the top differentially expressed genes) of genetic risk for binge-like ethanol drinking (85). These molecular signatures of risk were used to query the library of integrated network-based cellular signatures (LINCS)-1000 connectivity map to identify compounds that have the opposite effects on gene expression to the effects of selection for drinking ethanol to intoxication. Two compounds, pergolide and terreic acid, were identified and prioritized for in vivo testing. Although the molecular signature was derived from males, both compounds were able to successfully reduce binge-like drinking and BECs in male and female HDID mice. The success of this approach in both sexes may be due to: 1) targeting gene expression signatures rather than a single gene, 2) inclusion of gene expression data from several brain regions known to be involved in addiction, and 3) use of multiple prioritization methods to identify top candidate compounds in common from each prioritization approach.
In a related study, the transcriptome from the NAc of female HDID mice was used to identify the consequences of chronic binge-like ethanol drinking (42). These findings were contrasted with the effects of chemogenetically increasing neuronal activity in the NAc, a manipulation that results in lasting reductions in ethanol drinking, suggestive of adaptive neuroplasticity via transcriptional and epigenetic mechanisms. Genes were identified and prioritized for in vivo pharmacological testing based on three criteria: 1) expression levels altered by chronic binge level ethanol drinking that were ameliorated with chemogenetic stimulation, 2) genes over-represented in gene ontology analysis, and 3) genes considered to be hub genes. These criteria were applied to test whether the product of the Hdac4 hub gene could be targeted pharmacologically to reduce binge-like drinking. Administration of the HDAC4/5 inhibitor, LMK235, reduced chronic binge-like ethanol drinking in both male and female HDID-1 mice. Although this approach has not yielded a sex-specific outcome, it is important to consider the impact of sex at both the stage of molecular signature determination and testing for treatment efficacy. Please see supplemental information for approaches to interpreting sex differences in the transcriptome with integrative functional genomics.
CONCLUSIONS AND FUTURE DIRECTIONS
We have provided snapshots of how differences in the transcriptome align with differences between males and females in ethanol-associated behaviors. There are currently few studies comparing male and female transcriptomes, from the perspective of ethanol phenotypes. Despite the fact that multiple labs contributed to this paper, using a number of methodological and analytical approaches, sex differences in transcriptome findings relevant to alcohol effects and risk for use were consistently found, highlighting the importance of including sex as a biological factor in genomic investigations. Optimistically, what will emerge as these studies multiply are novel, sex-dependent therapeutic strategies for AUD, driven by the genomic and circuitry information, the latter for which sex-specific data particularly are sparse. Our knowledge is also incomplete with regard to cell types that respond to ethanol in males and females, as these too may be different. For example, Brenner et al. (86) found in a small sample of alcohol-dependent males and controls, post-mortem PFC cell type-specific effects of chronic alcohol use, primarily affecting astrocytes, oligodendrocytes and microglia. The latter observation aligns with the neuroimmune data noted above. The primary cilium is an underexplored feature of mental illness (87). Unique proteins expressed by the primary cilium (88), as well as a diverse array of signaling pathways (Figure 4), may emerge in treatment considerations, based on transcriptome and drug effect results pertinent to addiction and addiction risk (62,89,90). Of interest, Casanova Ferrer et al. (91) in the first meta-analysis of AUD and sex-specific effects on the post-mortem brain transcriptome, detected a cilium signal in females but not males.
Figure 4.
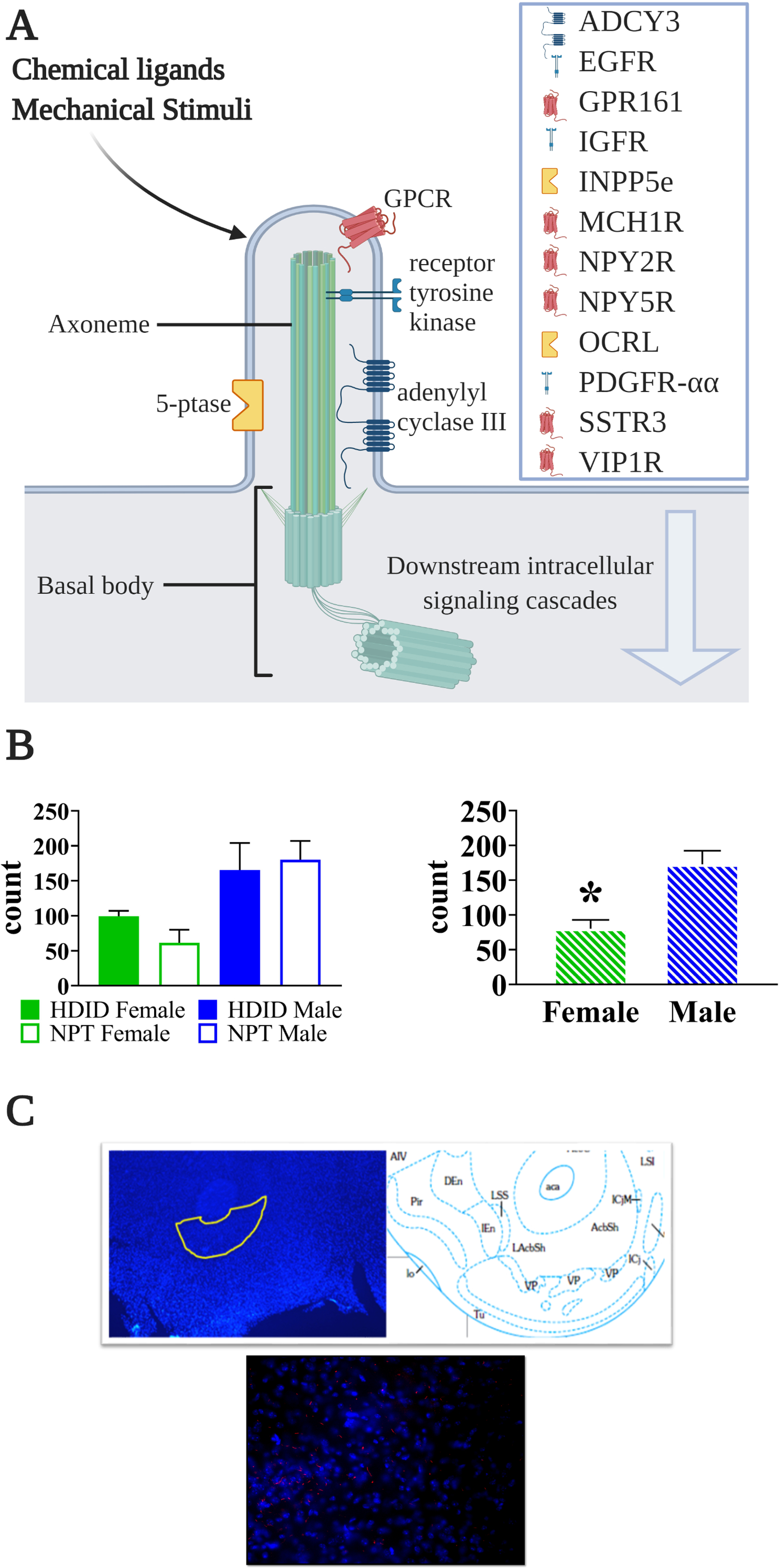
Primary cilia-specific protein (ADCY3) is expressed in greater quantity in the nucleus accumbens core in male than female mice. A) Schematic showing the protrusion of the cell membrane containing the primary cilium. Each cilium core is formed by an axoneme cytoskeletal structure and the cilium is enriched in the listed proteins, some of which are unique to the cilium. The basal body is the cellular organelle from which the cilia extends. Downstream intracellular signaling cascades include Ca2+, Wnt, phosphatidylinositols, and hedgehog, among others. Figure created with Biorender.com. B) Quantification of ADCY3 immunopositive cells counted from the nucleus accumbens core in high drinking in the dark (HDID) selected line mice and their NPT controls (n=3/sex/strain). Although the HDID mice did not differ from the control, male mice did show significantly higher ADCY3 immunoreactivity than female mice (p<0.005). C) Example of ADCY3 immunopositive cells in a nucleus accumbens section from a male mouse. 5-ptase, phosphoinositide 5-phosphatase; ADCY3, adenylate cyclase 3; EGFR, epithelial growth factor receptor; GPCR, G protein-coupled receptor; GPR161, G protein-coupled receptor161; IGFR, insulin-like growth factor receptor; INPP5e, inositol polyphosphate-5-phosphatase; MCH1R, melanin-concentrating hormone 1 receptor; NPY2R, neuropeptide Y2 receptor; NPY5R, neuropeptide Y5 receptor; OCRL, oculocerebrorenal syndrome of Lowe; PDGFR-αα, platelet-derived growth factor receptor α homodimer; SSTR3, somatostatin receptor isoform 3; VIP1R, vasoactive intestinal peptide receptor
Although we have emphasized that more data are needed to address sex differences in this research field, all reviews of sex difference data have to appreciate, and when possible account for, a community-wide ascertainment bias in publication: namely, that it is almost always easier to publish positive sex difference studies than negative counterpoints. The undesired risk is that the perception of sex differences gets serially magnified beyond the actual biological sex difference. One should therefore read the literature with the recognition of this inherent “sociology of science” bias. The best way to finesse this problem is to perform large mega and meta-analyses with data for which nearly all possible confounders have been eliminated. This, of course, is one of the advantages of animal research, which can choose to utilize isogenic males and females in controlled environments or can manufacture balanced heterogeneous stocks (e.g., (92)) for the study of sex x genotype x transcriptome (or circuitry) investigation.
Supplementary Material
ACKNOWLEDGEMENTS AND DISCLOSURES
Supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism grants P60AA010760, U01AA013484, R24AA020245, R24 AA13162, R01AA027096, R01 AA027096, R01AA 028680 and U01AA013519; the Andrews Genomics Fund; US Department of Veterans Affairs grants IK2BX002488,I01BX004699 and BX002966 and the VA Research Career Scientist Program.
Footnotes
The authors report no financial interest or potential conflicts of interest. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Robert Hitzemann, Department of Behavioral Neuroscience and Portland Alcohol Research Center, Oregon Health & Science University, Portland, Oregon.
Susan E. Bergeson, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas
Ari E. Berman, The Jackson Laboratory, Bar Harbor, Maine
Jason A. Bubier, The Jackson Laboratory, Bar Harbor, Maine
Elissa J. Chesler, The Jackson Laboratory, Bar Harbor, Maine
Deborah A. Finn, Veterans Affairs Portland Health Care System, Portland, Oregon
Lutz Hein, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas.
Paula Hoffman, Department of Pharmacology, University of Colorado, Aurora Colorado.
Andrew Holmes, Laboratory of Behavioral and Genomic Neuroscience, NIAAA.
Brent R. Kisby, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas
Denesa Lockwood, Department of Behavioral Neuroscience and Portland Alcohol Research Center, Oregon Health & Science University, Portland, Oregon.
Kerrie H. Lodowski, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas
Michelle McManus, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas.
Julie A. Owen, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas
Angela R. Ozburn, Department of Behavioral Neuroscience and Portland Alcohol Research Center, Oregon Health & Science University, Portland, Oregon Veterans Affairs Portland Health Care System, Portland, Oregon.
Praneetha Panthagani, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas.
Igor Ponomarev, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas.
Laura Saba, Department of Pharmaceutical Sciences, Skaggs School of Pharmacy & Pharmaceutical Sciences, University of Colorado Anschutz Medical Campus, Aurora Colorado.
Boris Tabakoff, Department of Pharmaceutical Sciences, Skaggs School of Pharmacy & Pharmaceutical Sciences, University of Colorado Anschutz Medical Campus, Aurora Colorado.
Aashlesha Walchale, Department of Pharmacology and Neuroscience, Texas Tech University Health Sciences Center, Lubbock, Texas.
Robert W. Williams, Department of Genetics, Genomics and Informatics, University of Tennessee Health Sciences Center, Memphis, Tennessee
Tamara J. Phillips, Department of Behavioral Neuroscience and Portland Alcohol Research Center, Oregon Health & Science University, Portland, Oregon
REFERENCES
- 1.Erol A, Karpyak VM (2015): Sex and gender-related differences in alcohol use and its consequences: Contemporary knowledge and future research considerations. Drug Alcohol Depend 156: 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Becker JB, Koob GF (2016): Sex Differences in Animal Models: Focus on Addiction. Pharmacol Rev 68: 242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilhelm CJ, Hashimoto JG, Roberts ML, Sonmez MK, Wiren KM (2014): Understanding the addiction cycle: a complex biology with distinct contributions of genotype vs. sex at each stage. Neuroscience 279: 168–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. (2019): Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10: 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richmond-Rakerd LS, Slutske WS, Lynskey MT, Agrawal A, Madden PAF, Bucholz KK, et al. (2016): Age at first use and later substance use disorder: Shared genetic and environmental pathways for nicotine, alcohol, and cannabis. J Abnorm Psychol 125: 946–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kendler KS, Edwards A, Myers J, Cho SB, Adkins A, Dick D (2015): The predictive power of family history measures of alcohol and drug problems and internalizing disorders in a college population. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet 168B: 337–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peltier MR, Verplaetse TL, Mineur YS, Petrakis IL, Cosgrove KP, Picciotto MR, McKee SA (2019): Sex differences in stress-related alcohol use. Neurobiol Stress 10: 100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpyak VM, Geske JR, Hall-Flavin DK, Loukianova LL, Schneekloth TD, Skime MK, et al. (2019): Sex-specific association of depressive disorder and transient emotional states with alcohol consumption in male and female alcoholics. Drug Alcohol Depend 196: 31–39. [DOI] [PubMed] [Google Scholar]
- 9.McHugh RK, Weiss RD (2019): Alcohol Use Disorder and Depressive Disorders. Alcohol Res Curr Rev 40. 10.35946/arcr.v40.1.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, et al. (2017): Prevalence of 12-Month Alcohol Use, High-Risk Drinking, and DSM-IV Alcohol Use Disorder in the United States, 2001–2002 to 2012–2013: Results From the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry 74: 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agabio R, Pisanu C, Gessa GL, Franconi F (2017): Sex Differences in Alcohol Use Disorder. Curr Med Chem 24: 2661–2670. [DOI] [PubMed] [Google Scholar]
- 12.Rehm J, Manthey J, Shield KD, Ferreira-Borges C (2019): Trends in substance use and in the attributable burden of disease and mortality in the WHO European Region, 2010–16. Eur J Public Health 29: 723–728. [DOI] [PubMed] [Google Scholar]
- 13.Cui C, Shurtleff D, Harris RA (2014): Neuroimmune mechanisms of alcohol and drug addiction. Int Rev Neurobiol 118: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson EK, Grantham EK, Warden AS, Harris RA (2019): Neuroimmune signaling in alcohol use disorder. Pharmacol Biochem Behav 177: 34–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crews FT, Lawrimore CJ, Walter TJ, Coleman LG (2017): The role of neuroimmune signaling in alcoholism. Neuropharmacology 122: 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hommer D, Momenan R, Kaiser E, Rawlings R (2001): Evidence for a gender-related effect of alcoholism on brain volumes. Am J Psychiatry 158: 198–204. [DOI] [PubMed] [Google Scholar]
- 17.Mogos MF, Salemi JL, Phillips SA, Piano MR (2019): Contemporary Appraisal of Sex Differences in Prevalence, Correlates, and Outcomes of Alcoholic Cardiomyopathy. Alcohol Alcohol Oxf Oxfs 54: 386–395. [DOI] [PubMed] [Google Scholar]
- 18.Briasoulis A, Agarwal V, Messerli FH (2012): Alcohol consumption and the risk of hypertension in men and women: a systematic review and meta-analysis. J Clin Hypertens Greenwich Conn 14: 792–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfefferbaum A, Rosenbloom M, Rohlfing T, Sullivan EV (2009): Degradation of association and projection white matter systems in alcoholism detected with quantitative fiber tracking. Biol Psychiatry 65: 680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehm J, Taylor B, Mohapatra S, Irving H, Baliunas D, Patra J, Roerecke M (2010): Alcohol as a risk factor for liver cirrhosis: a systematic review and meta-analysis. Drug Alcohol Rev 29: 437–445. [DOI] [PubMed] [Google Scholar]
- 21.Deak JD, Miller AP, Gizer IR (2019): Genetics of alcohol use disorder: a review. Curr Opin Psychol 27: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescott CA, Aggen SH, Kendler KS (1999): Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol Clin Exp Res 23: 1136–1144. [DOI] [PubMed] [Google Scholar]
- 23.Kendler KS, Ohlsson H, Sundquist J, Sundquist K (2018): Transmission of alcohol use disorder across three generations: a Swedish National Study. Psychol Med 48: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nestler EJ, Hyman SE (2010): Animal models of neuropsychiatric disorders. Nat Neurosci 13: 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadeau JH, Auwerx J (2019): The virtuous cycle of human genetics and mouse models in drug discovery. Nat Rev Drug Discov 18: 255–272. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster FE, Spiegel KS (1992): Sex differences in pattern of drinking. Alcohol Fayettev N 9: 415–420. [DOI] [PubMed] [Google Scholar]
- 27.Belknap JK, Crabbe JC, Young ER (1993): Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 112: 503–510. [DOI] [PubMed] [Google Scholar]
- 28.McKinzie DL, Nowak KL, Murphy JM, Li TK, Lumeng L, McBride WJ (1998): Development of alcohol drinking behavior in rat lines selectively bred for divergent alcohol preference. Alcohol Clin Exp Res 22: 1584–1590. [PubMed] [Google Scholar]
- 29.Vengeliene V, Vollmayr B, Henn FA, Spanagel R (2005): Voluntary alcohol intake in two rat lines selectively bred for learned helpless and non-helpless behavior. Psychopharmacology (Berl) 178: 125–132. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA (2008): Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol Fayettev N 42: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sneddon EA, White RD, Radke AK (2019): Sex Differences in Binge-Like and Aversion-Resistant Alcohol Drinking in C57BL/6J Mice. Alcohol Clin Exp Res 43: 243–249. [DOI] [PubMed] [Google Scholar]
- 32.Morales M, McGinnis MM, McCool BA (2015): Chronic ethanol exposure increases voluntary home cage intake in adult male, but not female, Long-Evans rats. Pharmacol Biochem Behav 139: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jury NJ, DiBerto JF, Kash TL, Holmes A (2017): Sex differences in the behavioral sequelae of chronic ethanol exposure. Alcohol Fayettev N 58: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varlinskaya EI, Spear LP (2004): Changes in sensitivity to ethanol-induced social facilitation and social inhibition from early to late adolescence. Ann N Y Acad Sci 1021: 459–461. [DOI] [PubMed] [Google Scholar]
- 35.Lynch WJ, Roth ME, Carroll ME (2002): Biological basis of sex differences in drug abuse: preclinical and clinical studies. Psychopharmacology (Berl) 164: 121–137. [DOI] [PubMed] [Google Scholar]
- 36.Schramm-Sapyta NL, Francis R, MacDonald A, Keistler C, O’Neill L, Kuhn CM (2014): Effect of sex on ethanol consumption and conditioned taste aversion in adolescent and adult rats. Psychopharmacology (Berl) 231: 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hager R, Lu L, Rosen GD, Williams RW (2012): Genetic architecture supports mosaic brain evolution and independent brain-body size regulation. Nat Commun 3: 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarthy MM, Nugent BM, Lenz KM (2017): Neuroimmunology and neuroepigenetics in the establishment of sex differences in the brain. Nat Rev Neurosci 18: 471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy MM (2020): A new view of sexual differentiation of mammalian brain. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 206: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorski RA, Gordon JH, Shryne JE, Southam AM (1978): Evidence for a morphological sex difference within the medial preoptic area of the rat brain. Brain Res 148: 333–346. [DOI] [PubMed] [Google Scholar]
- 41.Logrip ML, Milivojevic V, Bertholomey ML, Torregrossa MM (2018): Sexual dimorphism in the neural impact of stress and alcohol. Alcohol Fayettev N 72: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pozhidayeva DY, Farris SP, Goeke CM, Firsick EJ, Townsley KG, Guizzetti M, Ozburn AR (2020): Chronic Chemogenetic Stimulation of the Nucleus Accumbens Produces Lasting Reductions in Binge Drinking and Ameliorates Alcohol-Related Morphological and Transcriptional Changes. Brain Sci 10. 10.3390/brainsci10020109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purohit K, Parekh PK, Kern J, Logan RW, Liu Z, Huang Y, et al. (2018): Pharmacogenetic Manipulation of the Nucleus Accumbens Alters Binge-Like Alcohol Drinking in Mice. Alcohol Clin Exp Res 42: 879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cassataro D, Bergfeldt D, Malekian C, Van Snellenberg JX, Thanos PK, Fishell G, Sjulson L (2014): Reverse pharmacogenetic modulation of the nucleus accumbens reduces ethanol consumption in a limited access paradigm. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 39: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Logrip ML, Oleata C, Roberto M (2017): Sex differences in responses of the basolateral-central amygdala circuit to alcohol, corticosterone and their interaction. Neuropharmacology 114: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirson D, Oleata CS, Parsons LH, Ciccocioppo R, Roberto M (2018): CB1 and ethanol effects on glutamatergic transmission in the central amygdala of male and female msP and Wistar rats. Addict Biol 23: 676–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales M, McGinnis MM, Robinson SL, Chappell AM, McCool BA (2018): Chronic Intermittent Ethanol Exposure Modulation of Glutamatergic Neurotransmission in Rat Lateral/Basolateral Amygdala is Duration-, Input-, and Sex-Dependent. Neuroscience 371: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joffe ME, Winder DG, Conn PJ (2020): Contrasting sex-dependent adaptations to synaptic physiology and membrane properties of prefrontal cortex interneuron subtypes in a mouse model of binge drinking. Neuropharmacology 108126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prendergast BJ, Onishi KG, Zucker I (2014): Female mice liberated for inclusion in neuroscience and biomedical research. Neurosci Biobehav Rev 40: 1–5. [DOI] [PubMed] [Google Scholar]
- 50.Forger NG, Morin LP (1982): Reproductive state modulates ethanol intake in rats: effects of ovariectomy, ethanol concentration, estrous cycle and pregnancy. Pharmacol Biochem Behav 17: 323–331. [DOI] [PubMed] [Google Scholar]
- 51.Priddy BM, Carmack SA, Thomas LC, Vendruscolo JCM, Koob GF, Vendruscolo LF (2017): Sex, strain, and estrous cycle influences on alcohol drinking in rats. Pharmacol Biochem Behav 152: 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ford MM, Eldridge JC, Samson HH (2002): Microanalysis of ethanol self-administration: estrous cycle phase-related changes in consumption patterns. Alcohol Clin Exp Res 26: 635–643. [PubMed] [Google Scholar]
- 53.DiCarlo LM, Vied C, Nowakowski RS (2017): The stability of the transcriptome during the estrous cycle in four regions of the mouse brain. J Comp Neurol 525: 3360–3387. [DOI] [PubMed] [Google Scholar]
- 54.Duclot F, Kabbaj M (2015): The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol 16: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, et al. (1998): High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome Off J Int Mamm Genome Soc 9: 983–990. [DOI] [PubMed] [Google Scholar]
- 56.Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, et al. (2006): Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A 103: 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agrawal RG, Owen JA, Levin PS, Hewetson A, Berman AE, Franklin SR, et al. (2014): Bioinformatics analyses reveal age-specific neuroimmune modulation as a target for treatment of high ethanol drinking. Alcohol Clin Exp Res 38: 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B (2019): WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47: W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iancu OD, Colville AM, Wilmot B, Searles R, Darakjian P, Zheng C, et al. (2018): Gender-Specific Effects of Selection for Drinking in the Dark on the Network Roles of Coding and Noncoding RNAs. Alcohol Clin Exp Res. 10.1111/acer.13777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barkley-Levenson AM, Crabbe JC (2015): Genotypic and sex differences in anxiety-like behavior and alcohol-induced anxiolysis in High Drinking in the Dark selected mice. Alcohol Fayettev N 49: 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, McWeeney S (2012): Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics 28: 1592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hitzemann R, Phillips TJ, Lockwood DR, Darakjian P, Searles RP (2020): Phenotypic and gene expression features associated with variation in chronic ethanol consumption in heterogeneous stock collaborative cross mice. Genomics. 10.1016/j.ygeno.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dhaher R, Finn D, Snelling C, Hitzemann R (2008): Lesions of the extended amygdala in C57BL/6J mice do not block the intermittent ethanol vapor-induced increase in ethanol consumption. Alcohol Clin Exp Res 32: 197–208. [DOI] [PubMed] [Google Scholar]
- 64.Zhang B, Horvath S (2005): A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17. [DOI] [PubMed] [Google Scholar]
- 65.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, et al. (2008): A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci Off J Soc Neurosci 28: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. (2016): Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. (2013): Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tripathi PP, Di Giovannantonio LG, Sanguinetti E, Acampora D, Allegra M, Caleo M, et al. (2014): Increased dopaminergic innervation in the brain of conditional mutant mice overexpressing Otx2: effects on locomotor behavior and seizure susceptibility. Neuroscience 261: 173–183. [DOI] [PubMed] [Google Scholar]
- 69.Sherf O, Nashelsky Zolotov L, Liser K, Tilleman H, Jovanovic VM, Zega K, et al. (2015): Otx2 Requires Lmx1b to Control the Development of Mesodiencephalic Dopaminergic Neurons. PloS One 10: e0139697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Planques A, Oliveira Moreira V, Dubreuil C, Prochiantz A, Di Nardo AA (2019): OTX2 Signals from the Choroid Plexus to Regulate Adult Neurogenesis. eNeuro 6. 10.1523/ENEURO.0262-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spatazza J, Lee HHC, Di Nardo AA, Tibaldi L, Joliot A, Hensch TK, Prochiantz A (2013): Choroid-plexus-derived Otx2 homeoprotein constrains adult cortical plasticity. Cell Rep 3: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwarz JM, Bilbo SD (2012): Sex, glia, and development: interactions in health and disease. Horm Behav 62: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mayfield J, Harris RA (2017): The Neuroimmune Basis of Excessive Alcohol Consumption. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 42: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rath M, Guergues J, Pinho JPC, Zhang P, Nguyen TG, MacFadyen KA, et al. (2020): Chronic Voluntary Binge Ethanol Consumption Causes Sex-specific Differences in Microglial Signaling Pathways and Withdrawal-associated Behaviors in Mice. Alcohol Clin Exp Res. 10.1111/acer.14420 [DOI] [PubMed] [Google Scholar]
- 75.Blednov YA, Benavidez JM, Geil C, Perra S, Morikawa H, Harris RA (2011): Activation of inflammatory signaling by lipopolysaccharide produces a prolonged increase of voluntary alcohol intake in mice. Brain Behav Immun 25 Suppl 1: S92–S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warden AS, Azzam M, DaCosta A, Mason S, Blednov YA, Messing RO, et al. (2019): Toll-like receptor 3 activation increases voluntary alcohol intake in C57BL/6J male mice. Brain Behav Immun 77: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warden AS, Azzam M, DaCosta A, Mason S, Blednov YA, Messing RO, et al. (2019): Toll-like receptor 3 dynamics in female C57BL/6J mice: Regulation of alcohol intake. Brain Behav Immun 77: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grantham EK, Warden AS, McCarthy GS, DaCosta A, Mason S, Blednov Y, et al. (2020): Role of toll-like receptor 7 (TLR7) in voluntary alcohol consumption. Brain Behav Immun. 10.1016/j.bbi.2020.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, et al. (2015): Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86: 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Warden AS, Triplett TA, Lyu A, Grantham EK, Azzam MM, DaCosta A, et al. (2020): Microglia depletion and alcohol: Transcriptome and behavioral profiles. Addict Biol e12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finn DA, Hashimoto JG, Cozzoli DK, Helms ML, Nipper MA, Kaufman MN, et al. (2018): Binge Ethanol Drinking Produces Sexually Divergent and Distinct Changes in Nucleus Accumbens Signaling Cascades and Pathways in Adult C57BL/6J Mice. Front Genet 9: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayfield J, Ferguson L, Harris RA (2013): Neuroimmune signaling: a key component of alcohol abuse. Curr Opin Neurobiol 23: 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hashimoto JG, Wiren KM (2008): Neurotoxic consequences of chronic alcohol withdrawal: expression profiling reveals importance of gender over withdrawal severity. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 33: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crews FT, Sarkar DK, Qin L, Zou J, Boyadjieva N, Vetreno RP (2015): Neuroimmune Function and the Consequences of Alcohol Exposure. Alcohol Res Curr Rev 37: 331–341, 344–351. [PMC free article] [PubMed] [Google Scholar]
- 85.Ferguson LB, Ozburn AR, Ponomarev I, Metten P, Reilly M, Crabbe JC, et al. (2018): Genome-Wide Expression Profiles Drive Discovery of Novel Compounds that Reduce Binge Drinking in Mice. Neuropsychopharmacol Off Publ Am Coll Neuropsychopharmacol 43: 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brenner E, Tiwari GR, Kapoor M, Liu Y, Brock A, Mayfield RD (2020): Single cell transcriptome profiling of the human alcohol-dependent brain. Hum Mol Genet 29: 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pruski M, Lang B (2019): Primary Cilia-An Underexplored Topic in Major Mental Illness. Front Psychiatry 10: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phua SC, Lin Y-C, Inoue T (2015): An intelligent nano-antenna: Primary cilium harnesses TRP channels to decode polymodal stimuli. Cell Calcium 58: 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Bruin NMWJ, McCreary AC, van Loevezijn A, de Vries TJ, Venhorst J, van Drimmelen M, Kruse CG (2013): A novel highly selective 5-HT6 receptor antagonist attenuates ethanol and nicotine seeking but does not affect inhibitory response control in Wistar rats. Behav Brain Res 236: 157–165. [DOI] [PubMed] [Google Scholar]
- 90.Shiwaku H, Umino A, Umino M, Nishikawa T (2017): Phencyclidine-induced dysregulation of primary cilia in the rodent brain. Brain Res 1674: 62–69. [DOI] [PubMed] [Google Scholar]
- 91.Casanova Ferrer F, Pascual M, Hidalgo MR, Malmierca-Merlo P, Guerri C, García-García F (2020): Unveiling Sex-Based Differences in the Effects of Alcohol Abuse: A Comprehensive Functional Meta-Analysis of Transcriptomic Studies. Genes 11. 10.3390/genes11091106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagley JR, Chesler EJ, Philip VM, Center for the Systems Genetics of Addiction, Jentsch JD (2021): Heritability of Ethanol Consumption and Pharmacokinetics in a Genetically Diverse Panel of Collaborative Cross Mouse Strains and Their Inbred Founders. Alcohol Clin Exp Res. 10.1111/acer.14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


