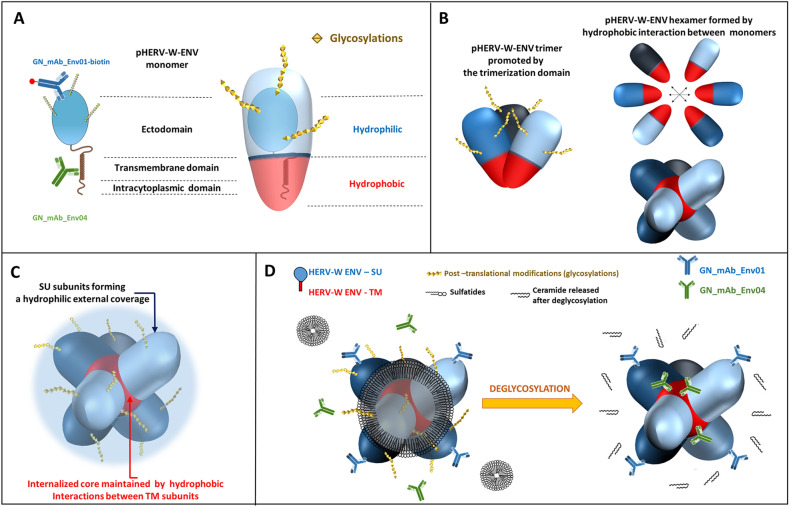
Fig. 5.
Schematic models for pHERV-W ENV antigens. A Schematic representation of pHERV-W ENV monomer with GN_mAb_Env01 and GN_mAb_Env04 antibodies binding sites. Blue and red overlays represent hydrophilic and hydrophobic domains of pHERV-W ENV, respectively. Yellow stars represent glycosylations. B pHERV-W ENV trimer formation driven by retroviral envelope trimerization domain conserved on pHERV-W ENV. pHERV-W ENV hydrophobic domains are still exposed despite trimer organization and retain the ability to induce hydrophobic interactions for hexameric formation, which does not reveal accurate but driven by monomers, as results showed. C pHERV-W ENV hexamer 3D-like representation. D Macrostructural representation showing that GN_mAb_Env01 epitope is accessible on the exposed hydrophilic ectodomain, contrary to GN_mAb_Env04 epitope buried in the hydrophobic core and masked by a layer of sulfatides. Deglycosylation induces sulfatides removal (by oside heads degradation) in addition of the opening of the deglycosylated protein structure, thereby making GN_mAb_Env04 epitope accessible to the antibody.