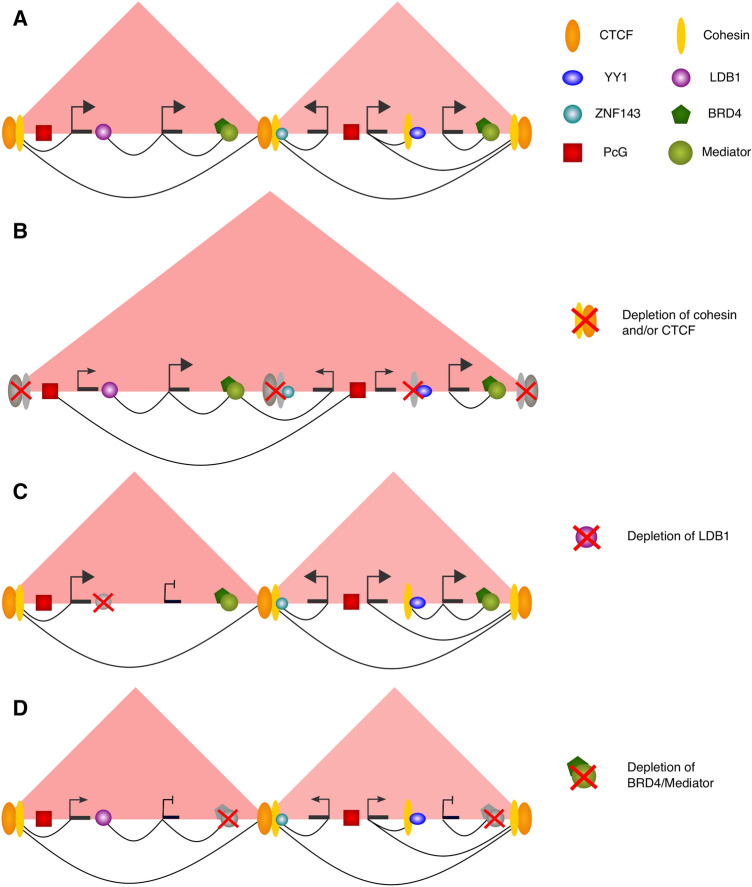
Fig. 2.
Effect of depletion of selected proteins on enhancer–promoter looping and gene expression. A In the wild type, enhancer–promoter contacts (arcs) occur in the context of large-scale contact domains such as TADs (red triangles) facilitated by the joint action of cohesin and CTCF (see “Large-scale chromosomal architecture”). Other cofactors relevant for enhancer–promoter communication (represented in various colours and shapes) can bind independently or in association with CTCF/cohesin at enhancers. Many of these factors are also found at promoters (not shown for simplicity). B Removal of CTCF or cohesin abolishes TADs. Long-range enhancer–promoter contacts proximal to TAD boundaries are disrupted, although some short-range contacts remain. In contrast, some long-range Polycomb-associated contacts (arc connecting red squares) and short-range enhancer–promoter contacts appear that spread across the native TAD boundaries. The transcriptional effects of this perturbation, however, remain relatively mild. C Removal of LDB1 abolishes an enhancer–promoter loop and leads to decreased gene expression for the affected gene. D Removal of BRD4 and/or Mediator does not disrupt enhancer–promoter contacts, but decreases gene expression. See section “Factors mediating enhancer–promoter looping” for details on individual factors and references to primary studies