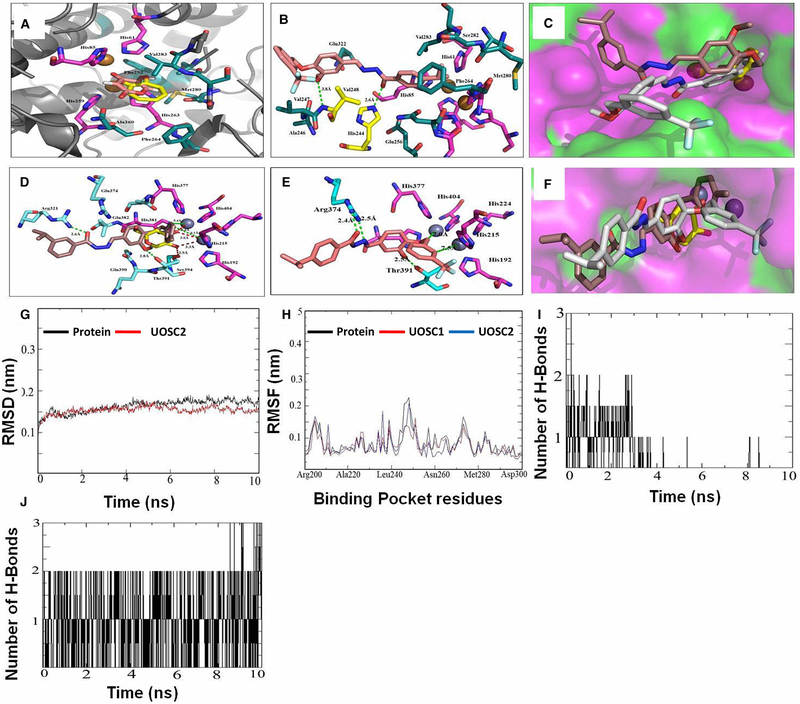
Figure 8. Computational modeling of UOSC-1, 2, 13, and 14 with fungal tyrosinase compared with human tyrosinase.
(A–C) Interaction of the native ligands (tropolone and kojic acid) and compounds UOSC1 or UOSC-2 within the active site of mushroom tyrosinase. (A) Illustrated the interaction of native ligand tropolone (yellow stick) and kojic acid (light pink stick) within the active site of mushroom tyrosinase (PDB:2Y9X). Copper ions represented as copper sphere surrounded with histidine residues (pink stick). (B) Illustrated the interactions of UOSC-2 (light brown stick) with residues (dark green stick). UOSC-2 showed H-bonds (yellow stick) with His244 and Val248. (C) Binding mode of native ligand, tropolone (yellow stick), UOSC-2 (white stick), and UOSC-1 (brown stick). UOSC-2 occupied the active center similar to native ligand. UOSC-2 is so near to copper ions at a distance of 3.8 Å. The remaining structure of UOSC-2 extended in the binding pocket, the surface represented as hydrophilic (pink), hydrophobic (green), and neutral (white). (D–F) Interactions of UOSC-1 or UOSC-2 and the native ligand, kojic acid within the active site of human tyrosinase. (D) Illustrated the interactions of kojic acid (light yellow stick) and UOSC-1 (brown stick) within the human tyrosinase active site (PDB:5M8M). (E) Illustrated the interactions of UOSC-2 (light brown stick) within human tyrosinase active site. (F) Illustrated the binding mode of UOSC-2 (white stick), UOSC-1 (brown stick), and kojic acid (yellow stick). Human tyrosinase surface represented as hydrophilic (pink), hydrophobic (green), and neutral (white). Zinc ions represented as violet spheres, histidines ligands represented as pink stick and pocket residues are represented as cyan stick. Kojic acid, the native ligand represented as yellow stick. H-bonds between UOSC-2 and the active site are green dots, while H-bonds between kojic acid and the pocket site are red dots. H-bonds between kojic acids and Zn ions are shorter than the H-bonds between UOSC-2 and Zn ions. The H-bonds between UOSC-2 and Zn ions are not in accepted H-bond length. (G–J) Molecular Dynamics (MD) simulation of the newly designed tyrosinase inhibitors. (G) Illustrated the RMSD values over 10 ns simulations of the protein (black) and UOSC-2 (red). (H) Illustrated the RMSF values in nanometer over 10 ns simulations of the protein (black) and UOSC-2 (blue). (I–J) H-bonds interaction over 10 ns MD simulation of (I) kojic acid and (J) UOSC-2.