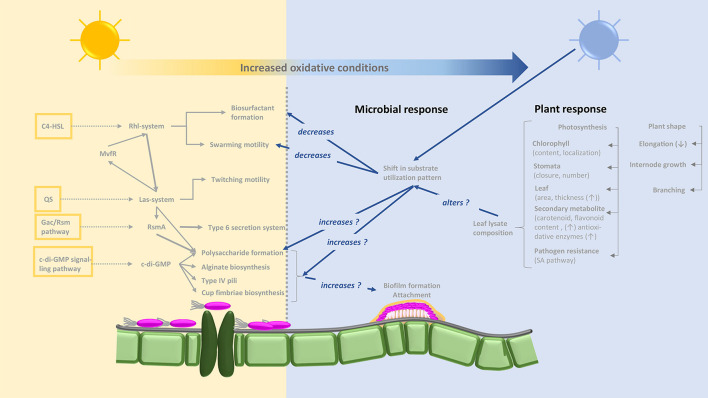
FIGURE 8.
Concept-based map illustrating putative interactions between light regime (non-blue light: yellow background; blue light: blue background), plant responses and responses of Pseudomonas sp. based on the results obtained and including literature knowledge. Pathways for biosurfactant and biofilm formation are based on Kyoto Encyclopedia of Genes and Genomes (KEGG) map02025 “Biofilm formation–Pseudomonas aeruginosa” (http://www.kegg.jp/kegg-bin/highlight_pathway?scale=1.0&map=map02025&keyword=biofilmPseudomonas). The yellow framed boxes on the left display pathways involved into biofilm formation and the dotted arrows link to the main signaling cascades. The C4-HSL = N-butyryl-l-homoserine lactone, links to the Rhl-system = two lux-type quorum sensing systems, and further to the Las-system. In Pseudomonas aeruginosa this is consisting of two components, namely an autoinducer synthesizing acyl homoserine lactone (AHL) from methionine and a transcriptional controller and mediates the formation of C4-HSL (N-butanoyl homoserine) which enables communication between bacterial cells among others biofilm formation. Interlinked between these the MvfR = transcriptional regulator moderating the expression of QS-based virulence factors, mediates a feedback system. The QS, quorum sensing, also acts into the Las-system impacting together with the Gac, global regulatory system for activation of antibiotic and cyanide synthesis, the RmsA, small RNA binding protein; the Gac/Rms pathway allows the organism to switch between primary and secondary metabolism; c-di-GMP, Bis-(3′–5′)-cyclic dimeric guanosine monophosphate, then also majorly acts on processes of the biofilm formation via impacting features like Polysaccharide formation, alginate biosynthesis, etc. According to the experimental findings, it is clear, that blue light regimes impacts several features of bacterial habit in the indicated way. However, it needs further investigation to identify the molecular mechanisms and locations of these influences.