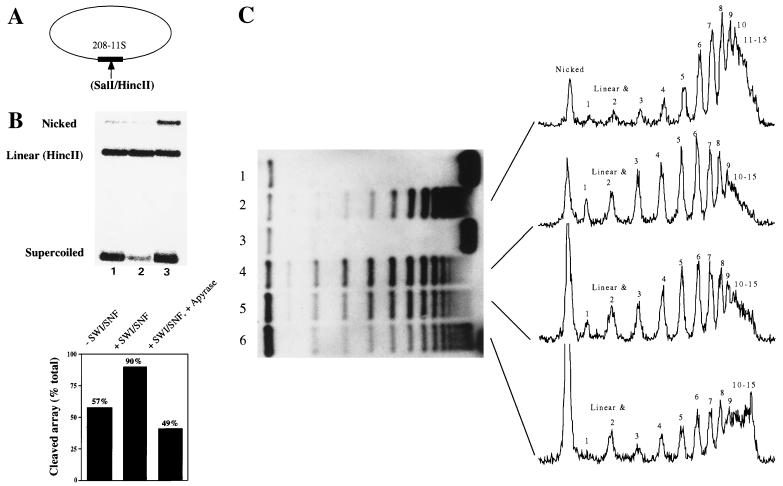
FIG. 2.
Remodeled nucleosomes do not accumulate on closed circular nucleosomal arrays. (A) Nucleosomal arrays were reconstituted on negatively supercoiled closed circular plasmids bearing a unique SalI/HincII site on a single 5S nucleosome positioning sequence. (B) The closed circular arrays were subjected to HincII digestion for 30 min followed by a 30-min incubation with (lane 2) or without (lane 1) SWI-SNF. Alternatively, the arrays were incubated for 30 min with SWI-SNF, followed by removal of ATP with apyrase and digestion with HincII for 30 min (lane 3). (C) A supercoiling assay was performed on the closed circular nucleosomal arrays (lane 1) in order to detect remodeling as changes in plasmid linking number. Addition of calf thymus topoisomerase I resulted in the appearance of approximately 15 bands after deproteination and agarose gel electrophoresis (lane 2). Incubation of the arrays with SWI-SNF and ATP for 30 min had no effect on topology (lane 3). Addition of SWI-SNF, ATP, and topoisomerase I resulted in a redistribution of the topoisomers (lane 4). Removal of ATP by addition of apyrase after 30 min of incubation in the presence of SWI-SNF plus topoisomerase did not affect the distribution of topoisomers (lane 5). Incubation of the arrays with SWI-SNF and ATP for 30 min followed by apyrase and then topoisomerase resulted in a topoisomer distribution similar to that in lane 2 (lane 6). Quantification of the autoradiograph indicates that for lanes 2 and 6, the predominant band corresponds to a linking number of 8; for lanes 4 and 5, the predominant band corresponds to a linking number of 6. Note that apyrase treatment of the circular array results in an increase in the nicked plasmid form (B, lane 3; C, lanes 5 and 6). Apyrase-induced plasmid nicking does not affect HincII activity or the relative distribution of topoisomers upon topoisomerase I treatment of the array (data not shown).