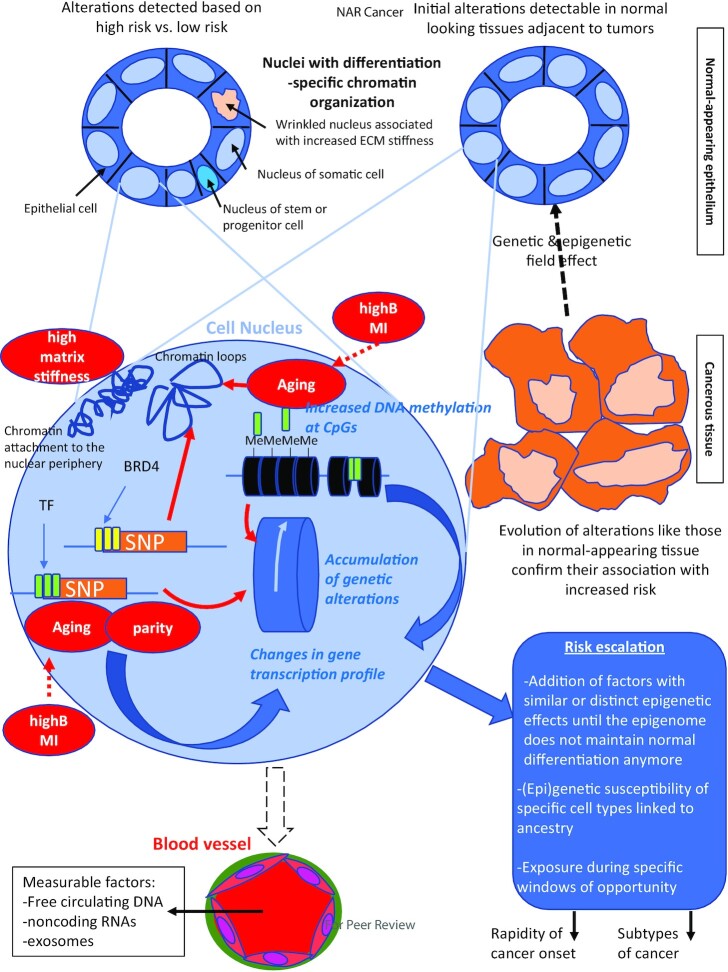
Figure 1.
Cancer risk escalation in the cell nucleus. It is essential to study changes in the epigenome and its related parameters that are associated with cancer risk in the normal tissue of origin. For the breast, the luminal epithelium is organized as a monolayer (internal to the layer of myoepithelial cells- not shown) within which cell heterogeneity is represented by different phenotypes and the presence of a small percentage of progenitor cells. These cells have a different organization of chromatin and its epigenetic content that might be detectable via a different shape and size of the nucleus. The source of this tissue might be from reduction mammoplasty (left) or from biopsies from normal-appearing tissue adjacent to tumors (right). The association of certain epigenetic alterations with risk is sometime confirmed because of an increase or an extension of these alterations in the same DNA regions as cancer develops and progresses. The breast epithelium may be reproduced in 3D cell culture to enable the mechanistic investigation of risk factors. The nucleus (pale blue magnified circle) of an epithelial cell at risk may display different types of chromatin alterations, like DNA methylation at CpG sites (often increased), changes in chromatin loops and anchorage/condensation. The presence of SNPs associated with risk at DNA regulatory regions (e.g. super enhancers marked by BRD4, TF binding regions) modifies gene transcription via a direct impact on ligand binding or through changes in chromatin looping. Depending on the risk factors (red bubbles) one or more effect on chromatin has been identified so far such as changes in DNA methylation, TF binding or expression, and chromatin compaction. Importantly, risk factors may also feed into others’ effects and could strengthen the extent of alterations as shown by the dashed red arrows. There are at least three possible measurable means (written in blue) to build up risk and that could help identify epigenome-related thresholds to cancer onset: DNA methylation profiles, genetic alterations and gene expression profiles. Moreover, SNPs and DNA methylation influence each other leading to the accumulation of genetic alterations. As illustrated in the ‘risk escalation’ box, how the risk is built in the chromatin is likely to influence the speed at which the epigenome loses control of normal differentiation and the resulting subtype of cancer. The blood stream is an important compartment to routinely measure cancer risk because of the presence of circulating DNA, ncRNAs and exosomes. Circulating DNA and ncRNAs might be specific or nonspecific indicators of risk, whereas exosomes might be traceable to a specific tissue at risk. The combination of different measurable parameters might be necessary to identify a risk of cancer in a specific tissue and how the risk was built (i.e., the epigenetic pathways altered, the type of cells involved) so that interventions may be tailored to the origin(s) of the increase in risk.