Figure 6.
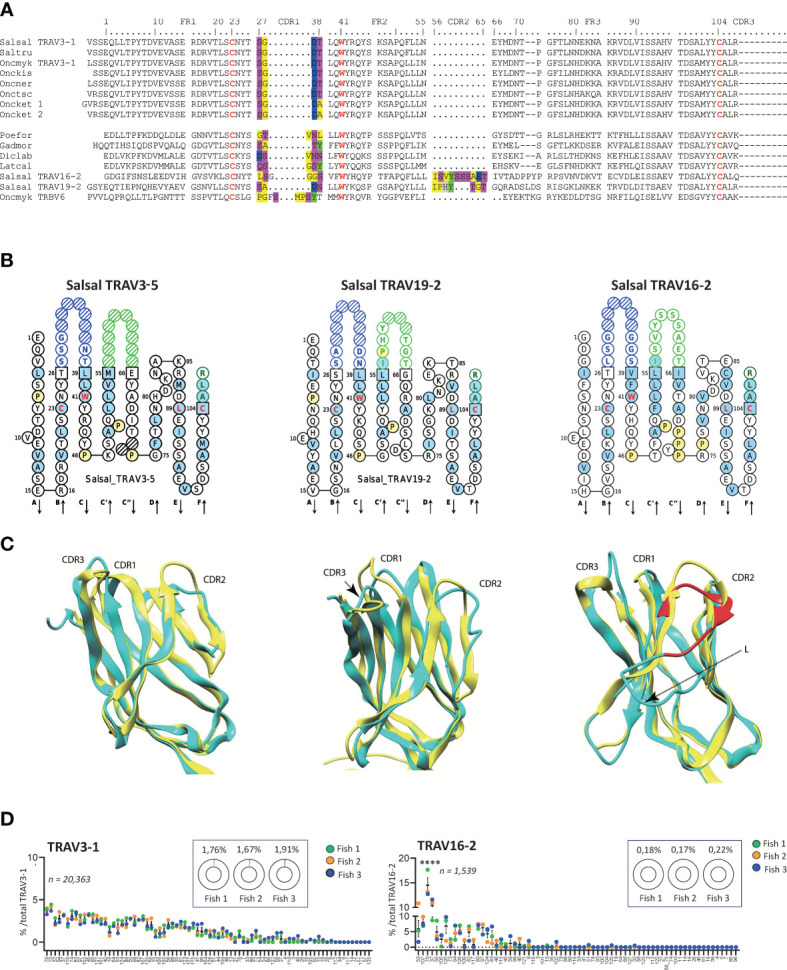
Variations of CDR2-IMGT across salmonid TRAV. (A) Multiple alignment of selected TRAV (TRAV22, TRAV16, TRAV19) protein sequences from Atlantic salmon (Salsal) and rainbow trout (Oncmyk) with sequences presenting similar features from other Salmonids and other teleosts. (B) IMGT Collier de Perles of Atlantic salmon TRAV3-5, TRAV19-2, TRAV16-2, illustrating CDR2-IMGT length variation. (C) Salmon TRAV3, TRAV19, and TRAV16 (cyan) aligned to human TR B7 alpha chain V domain. Note the lack of CDR2 in salmon TRAV3. Salmon TRAV19 is shown for comparison of CDR2 modelling, and TRAV16 domain for its very long CDR2 region (in red) that induces an additional loop (L) in the model, which may not reflect the reality but illustrates the constraint on the domain. (D) Relative TRAJ usage shown as percentage of total reads containing a specific TRAJ gene for TRAV3-1 or TRAV19-9 containing productive rearrangements. Individual TRAJ genes are shown on the X-axis, average number of reads from each fish containing the indicated TRAV-TRAJ combination are shown (fish 1 is in green, fish 2 in orange and fish 5 in blue). The total number of sequences for each TRAV is indicated and the inlaid box shows the average representation of each TRAV per total number of productive rearrangements from each fish. Atlantic salmon, Salsal; rainbow trout, Oncmyk; Brown trout, Saltru; Coho, Onckis; Chinook salmon, Onctsc; Chum salmon, Oncket; Sockeye, Oncner; African molly, Poefor; Atlantic cod, Gadmor; barramundi perch, Latcal).
