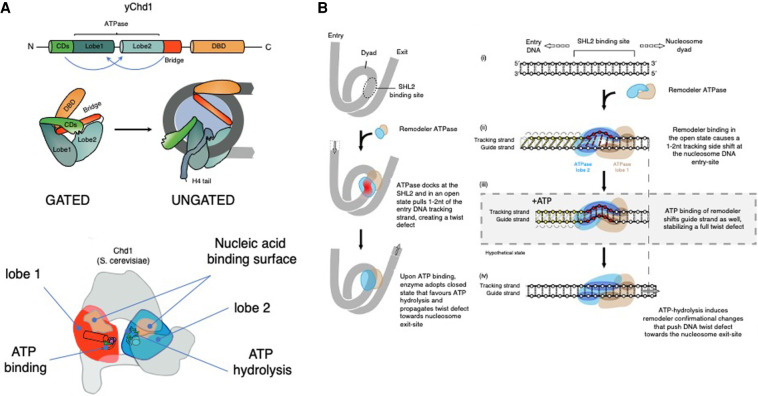
Figure 2.
Autoinhibitory regulation and nucleosome sliding mechanism of SF2-type chromatin remodelers. (A) The ATPase motor of CHD enzymes is comprised of two RecA-like lobes ([red] lobe1, [blue] lobe2) that adopt a closed confirmation to form a contiguous surface that allows nucleic acid binding and efficient ATP hydrolysis. Biophysical and structural studies of yChd1, CHD2, and CHD4 revealed autoinhibitory domain organization whereby flanking domains fold onto the ATPase motor to prevent activation by improper substrates (Hauk et al. 2010; Ryan et al. 2011; Morra et al. 2012; Watson et al. 2012; Liu et al. 2014a). The N-terminal tandem chromodomains (CD; green) of yChd1 contain a linker helix with several conserved acidic residues that stacks onto the DNA-binding surface of lobe2 and negatively regulates yChd1 activity (Hauk and Bowman 2011; Narlikar et al. 2013). Full inhibition is achieved through folding of the “bridge” inhibitory domain adjacent to the ATPase motor (Bridge; orange). Moreover, autoinhibitory interaction of the ATPase motor with the C-terminal domains uncouples ATP hydrolysis from productive DNA translocation of Chd1p and CHD4, respectively (Hauk et al. 2010; Morra et al. 2012; Watson et al. 2012). Upon chromatin binding, interactions of CHD domains with linker DNA and protruding histone H4 tails (H4; gray) trigger major conformational change and dislodging of the autoinhibitory domains, allowing the ATPase motor to adopt an active conformation (Hauk and Bowman 2011; Nodelman et al. 2021). The interplay between the ATPase motor and flanking domains not only provides means for autoinhibition but also can promote substrate recognition and ATPase activity. In the case of CHD4, the N-terminal tandem chromodomains and PHD domains stimulate ATPase motor activity and DNA translocation (Morra et al. 2012; Watson et al. 2012), whereas the respective C-terminal DNA-binding regions of CHD1, CHD2, and CHD7 stimulate activity of the ATPase motor (Ryan and Owen-Hughes 2011; Bouazoune and Kingston 2012; Liu et al. 2014a). Thus, intradomain allosteric regulation keeps the enzyme in an inactive conformation to ensure substrate specificity, while on the other hand facilitating ATPase motor activity and DNA translocation. (GATED) yChd1 closed confirmation, (UNGATED) open confirmation, (light blue) nucleosome core, (gray) wrapped DNA. Features required for the efficient ATP binding and hydrolysis in lobe1 and lobe2 are indicated: ATP binding (magenta circles), ATP hydrolysis (green lines), and nucleic acid binding surfaces on each lobe (opaque areas) (Hauk and Bowman 2011). Top panel cartoon adapted with permission from Springer Nature from Mueller-Planitz et al. (2013), © 2013. Bottom panel republished with permission of Elsevier Science and Technology Journals from Hauk and Bowman (2011); permission conveyed through Copyright Clearance Center, Inc. (B) Historically, nucleosome crystal structure and earlier models of nucleosome sliding revealed that nucleosomal DNA can accommodate an extra base pair at the internal DNA location termed superhelix location 2 (SHL2) (Luger et al. 1997), which is a docking site for many ATP-dependent chromatin remodelers. Recent evidence shows that CHD, ISWI, and Snf2-type remodelers shift DNA discontinuously around the nucleosome (Farnung et al. 2017, 2020; Winger et al. 2018; Sabantsev et al. 2019; Yan and Chen 2020; Zhong et al. 2020; Nodelman et al. 2021). Remarkably, in an open conformation, the ATPase motor bound at the nucleosomal SHL2 position pulls 1–2 bp of DNA at the nucleosome entry site, creating a shift of only one DNA strand (tracking strand) with respect to the other (guide strand), resulting in a twist defect. Subsequent ATP binding induces closed confirmation of the ATPase motor, pulling the entire base pair of the DNA at SHL2 and forcing the DNA twist defect toward the nucleosomal exit site. Upon ATP hydrolysis, the motor readopts the open confirmation again, priming the nucleosome for the next translocation cycle. This sequential, ATP-driven conformational transition between open and closed states propels the DNA around the nucleosome histone core (Liu et al. 2017; Farnung et al. 2017, 2020; Armache et al. 2019; Nodelman et al. 2021; Yan and Chen 2020). Republished with permission of Elsevier Science and Technology Journals from Bowman (2019); permission conveyed through Copyright Clearance Center, Inc.