Abstract
Vocal learning, the ability to imitate sounds from conspecifics and the environment, is a key component of human spoken language and learned song in three independently evolved avian groups—oscine songbirds, parrots, and hummingbirds. Humans and each of these three bird clades exhibit specialized behavioral, neuroanatomical, and brain gene expression convergence related to vocal learning, speech, and song. To understand the evolutionary basis of vocal learning gene specializations and convergence, we searched for and identified accelerated genomic regions (ARs), a marker of positive selection, specific to vocal learning birds. We found avian vocal learner-specific ARs, and they were enriched in noncoding regions near genes with known speech functions or brain gene expression specializations in humans and vocal learning birds, including FOXP2, NEUROD6, ZEB2, and MEF2C, and near genes with major neurodevelopmental functions, including NR2F1, NRP2, and BCL11B. We also found enrichment near the SFARI class S genes associated with syndromic vocal communication forms of autism spectrum disorders. These findings reveal strong candidate noncoding regions near genes for the evolutionary adaptations that distinguish vocal learning species from their close vocal nonlearning relatives and provide further evidence of molecular convergence between birdsong and human spoken language.
Vocal learning is a critical component of spoken language in humans, but understanding of the molecular mechanisms underlying vocal learning and human speech development remains incomplete. Vocal learning evolved independently in humans and at least four nonhuman mammalian lineages (cetaceans, bats, elephants, and pinnipeds) and three avian lineages (oscine songbirds, parrots, and hummingbirds) (Petkov and Jarvis 2012). Vocal learning species share a number of characteristic traits not found in vocal nonlearning species including: critical periods for learned imitation of new sounds; infant babbling; deafness induced deterioration of vocalization; dialects (Doupe and Kuhl 1999; Bolhuis et al. 2010; Petkov and Jarvis 2012); specialized neural pathways that control the vocal organs (syrinx in birds, larynx in mammals) (Wild et al. 1997; Petkov and Jarvis 2012); and specialized gene expression in the brain regions comprising the neural pathways (Hara et al. 2012; Pfenning et al. 2014; Lovell et al. 2018). The shared neural pathways consist of a cortico-striato-thalamo-cortical loop essential for learning and a motor cortex direct projection from the forebrain vocal motor control regions to the brainstem vocal motor neurons (Fig. 1A; Doupe and Kuhl 1999; Bolhuis et al. 2010; Arriaga et al. 2012). These neural pathways are either absent in vocal nonlearning species or rudimentary in many species according to a continuum hypothesis of vocal learning (Wild et al. 1997; Arriaga et al. 2012; Petkov and Jarvis 2012; Liu et al. 2013; Pfenning et al. 2014; Jarvis 2019).
Figure 1.
Convergent lineages. (A) Diagram of the vocal learning–related brain regions and circuits in oscine songbirds and human. Both species are characterized by anterior learning (white arrows) and posterior production (solid black arrows) pathways, including a direct neuronal projection (red arrow) from the forebrain to the brain stem. (B) A phylogeny of the taxa in this study using the topology of Jarvis et al. (2014) with branch lengths inferred from fourfold degenerate sites. Branches are colored by convergent phenotype: Red: vocal learners; blue: waterbirds, which are used as a biological control for some analyses; black: species that do not belong to either of these groups. Abbreviations: Songbird brain: Area X, Area X of the striatum; AV, nucleus avalanche; DLM, dorsolateral nucleus of the medial thalamus; DM, dorsal medial nucleus of the midbrain; HVC, high vocal center; LMAN, lateral magnocellular nucleus of the anterior nidopallium; LMO, lateral oval nucleus of the mesopallium; Nif, interfacial nucleus of the nidopallium; RA, robust arcopallium; XII, bird twelfth nerve nucleus. Human brain: Am, nucleus ambiguous; ASt, anterior striatum; aT, anterior thalamus; LMC, laryngeal motor cortex; LSC, laryngeal somatosenosry cortex; PAG, periaqueductal gray.
This convergence of advanced vocal learning suggests that nonhuman vocal learning species may be used as models for the study of human speech disorders, which affect about 6% of children (Law et al. 2000). Early intervention in speech development disorders, including some autism spectrum disorders (ASDs), can improve patient outcomes (Dawson et al. 2010). Prenatal and early childhood genetic screening are therefore potentially powerful therapeutic tools, but they require known causal genetic mechanisms to screen. A more complete understanding of the genetic basis of vocal learning may provide valuable candidates for such screenings.
Genomic markers of positive selection can be a powerful guide to elucidating the molecular pathways underlying traits relevant to human phenotypes and diseases as demonstrated by recent studies of longevity (Keane et al. 2015), immunity (Zhang et al. 2013), and obesity (Liu et al. 2014). In humans, positively selected accelerated regions (ARs), characterized by DNA sequence conservation in nonhuman species and faster than neutral DNA sequence evolution in humans, are enriched for neurological functions, potentially related to our evolution of larger brains and language (Kamm et al. 2013; Oksenberg et al. 2013; Boyd et al. 2015).
Here, we conducted an AR analysis on a genome alignment of 33 bird species including 12 vocal learners (seven oscine songbirds, four parrots, and one hummingbird) and their nearest vocal nonlearning relatives (three suboscine songbirds, one falcon, one swift, and one nightjar) (Figure 1B; Supplemental Table S1). We identified ARs specific to one or more of the three vocal learning clades, identified candidate associated genes, and tested for convergent acceleration across lineages.
Results
Accelerated region discovery and quality assessment
We aligned all 33 draft avian genomes, representing most major clades of birds, vocal learners, and their nearest nonlearning relatives (Fig. 1B; Supplemental Table S1) using Progressive Cactus (commit 95f1c43c9740201aec52844c085cc3bb92fb5757) (Paten et al. 2011). We found that Cactus was able to align an average of ∼70% of the best annotated genome, the chicken, to most other species (28 of the remaining 32 species) (Fig. 2A,B). This is a more than twofold improvement over previous avian whole-genome comparative studies (Zhang et al. 2014) that recovered 31.6% of the chicken genome in the alignment with the same proportion of species.
Figure 2.
Alignment and AR distribution information. (A) The fraction of each species genome aligned to the chicken, the genome with the most complete annotation in this study. (B) The fraction of the chicken genome with at least N species aligned to chicken (including chicken). (C) Distributions of ARs among the vocal learning acceleration tests in this study. In all three clades, the majority of ARs are intergenic (purple).
In keeping with previous AR studies (Pollard et al. 2006; Hubisz et al. 2011), to identify ARs specific to vocal learning birds, we first identified evolutionarily conserved elements (ECEs) in vocal nonlearning birds using phastCons (most-conserved criterion; PHAST v1.4) (Siepel et al. 2005). After filtering regions with ambiguous orthology between chicken and vocal learners, we retained 276,412 ECEs of at least 100 bp, spanning a combined 51 Mb. We further excluded any conserved element with an insufficient number of aligned species both in total and in the clade being tested for acceleration (see Methods), as well as any conserved element exhibiting evidence of recombination driven by GC-biased gene conversion, which can create acceleration without positive selection (Katzman et al. 2010), in the lineage being tested for selection (Supplemental Fig. S1). We found that the GC-biased regions tended to be at the ends of the chromosomes (Supplemental Fig. S2), which are known to be enriched for recombination in birds (Groenen et al. 2009; Backström et al. 2010), indicating that the filtration is appropriate.
We tested these ECEs in vocal nonlearners for ARs in each of the vocal learning clades using phyloP ACC (PHAST v1.4) (Pollard et al. 2006), which tests whether the rate of nucleotide substitution in a foreground group (a vocal learning clade) is substantially greater than the background group (all nonlearners) (Siepel et al. 2005; Hubisz et al. 2011). As a biological control set, we tested for ARs in another convergent group of avian lineages, waterbirds (ducks [mallard, Anseriformes], flamingos [American flamingo, Phoenicopteriformes], plovers [killdeer, Charadrifromes], and core waterbirds [emperor penguin and crested ibis, Aequornithia]) following the same pipeline as above, by ascertaining ECEs in all nonwaterbirds, including vocal learners.
We found 3608 oscine songbird, 3400 parrot, and 1795 hummingbird ARs, as well as 858 mallard, 105 flamingo, 133 killdeer, and 147 core waterbird ARs (Supplemental Tables S2–S4). Most vocal learning bird ARs were noncoding (91%–95%), with the largest subset being intergenic, more than 10 kb from a gene body (51%–62%) (Fig. 2C). Rates of acceleration varied among ARs within lineages and to a lesser degree between lineages (Supplemental Fig. S3). The total number of accelerated regions observed varied 34-fold between the most AR-rich lineages (oscines) and the least AR-rich lineage (flamingo). To explore the reason behind this, we conducted additional acceleration tests on each terminal branch of the phylogeny leading to a nonlearner. We found a linear correlation (R2= 0.63) between the number of ARs and branch length, inferred from fourfold degenerate sites by phyloFit (PHAST v1.4) (Fig. 3; Siepel et al. 2005; Hubisz et al. 2011). This relationship suggests that selection acting in a clocklike fashion is primarily responsible for the variation in the number of ARs observed in tests for acceleration on a single branch of the phylogeny.
Figure 3.

Relationship between branch length as inferred from fourfold degenerate sites in our 33-way bird alignment and the number of ARs found in each nonlearning bird. There was a broadly linear relationship between branch length and number of ARs ascertained, regardless of lineage, consistent with a clocklike accumulation of ARs.
To test for a possible effect of also having greater numbers of species represented in a clade, we conducted down-sampling experiments in the vocal learning clade with the most species sequenced (oscine songbirds), which happens to also be the clade with the most bird species (5000 of the ∼10,500 species). We repeated the oscine clade AR analysis with one to seven species and identified the number of ARs observed per lineage. We found increasingly larger number of ARs with more species, although there was a decrease when moving from one to two species (Supplemental Fig. S4, black). This included emergence of ARs not identified when any of the lineages are tested individually (Supplemental Fig. S4, red), indicating that the inclusion of multiple species yields an increase in statistical power. As we added taxa from one to four oscines, we observed an increasing ability to identify ARs as accelerated, but statistical significance continued to increase as additional taxa were added beyond the fourth species (Supplemental Fig. S5A,B). This indicates that beyond the number of ARs detected, the pattern of selection on an individual conserved element tended to converge toward the full data set result as we added more species to the analysis.
To further investigate whether vocal learning bird ARs were driven by a single taxon or by selection relevant to all species in the clade, which we would predict to be more likely related to vocal learning, we tested for acceleration in each oscine songbird species using with phyloP -CONACC (Pollard et al. 2006). Only two ARs were the result of acceleration specific to a single oscine songbird species, and only five ARs were the result of acceleration in <50% of oscine songbirds (Supplemental Fig. S6; Supplemental Table S5). This gives us confidence that most of the ARs we identified result from selection impacting most or all of the tested clade.
We also tested the influence of nonlearning taxa on AR discovery. We hypothesized that near relatives of vocal learners would exert a greater influence on AR ascertainment than distant relatives. To test this hypothesis, we compared the removal of oscine songbirds near vocal nonlearning relatives (e.g., suboscines) with the removal of randomly selected neoaves, the clade comprising all species except chickens and ducks (chicken, greater prairie chicken, mallard) and ratites (ostrich and white throated tinamous). In both cases, we observed decreases in power, but the effect was much larger when removing near relatives of oscines (Supplemental Fig. S5C). For example, whereas removing five distantly related background species resulted in a loss of <5% of oscine songbird ARs, removing just the three suboscines resulted in a loss of >30% of oscine songbird ARs. This indicates that the inclusion of these close outgroup species is important, even though they were not directly tested for acceleration.
Some ARs may be the result of loss of constraint rather than positive selection. However, over the ∼30–100 million lineage divergence timespans being considered in this analysis (Jarvis et al. 2014), one or more episodes of positive selection or an overarching loss of constraint might produce similar signatures of acceleration. In either case, it would represent a probable deviation in the function of the conserved element so that even a loss of constraint would still be indicative a likely evolutionarily relevant event.
Accelerated regions in vocal learning birds
The distribution of both ARs and ECEs varied across the genome and include regions of high AR density, that is, acceleration hotspots, which in past studies have been found to be major targets of adaptive evolution (Booker et al. 2016). To ascertain whether the AR hotspots we found in this study are indeed areas of AR enrichment independent of ECE density, we tested for a correlation between AR and ECE density. Consistent with AR studies in humans (Pollard et al. 2006; Capra et al. 2013; Hubisz and Pollard 2014), we found that AR densities in vocal learning birds were not strongly correlated with the number of ECEs in a region (Supplemental Fig. S7). These findings suggest that ARs in vocal learners are not randomly distributed among ECEs in their genomes but in specific regions of the genome.
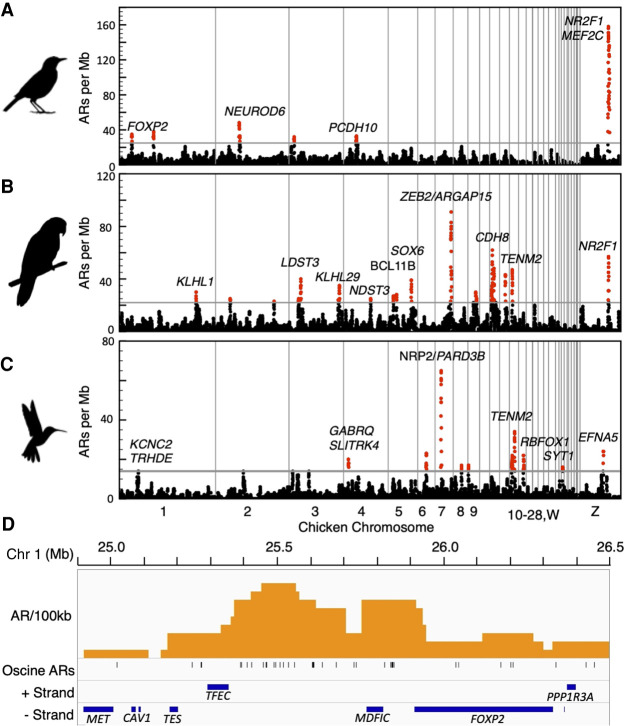
To identify potential acceleration hotspots in vocal learners, we randomly sampled ECEs equal to the number of ARs observed and calculated the genome-wide maximum density. AR densities in a lineage that exceeded the genome-wide maximum density in at least 95 of 100 (95%) resamplings were considered acceleration hotspots. We found between six and 14 AR hotspots in all three vocal learning bird lineages (Fig. 4A–C) as well as five less dense ones in the mallard but no AR hotspots in the other waterbirds (Supplemental Fig. S8A–D). Most AR hotspots were not found within gene bodies but in noncoding regions (Fig. 4D; Supplemental Figs. S9–S11). Some of the nearby genes to the AR hotspots of vocal learners included those with previously implicated roles in vocal learning. This notably included the transcription factors FOXP2, NEUROD6, and MEF2C among the six oscine AR hotspots (Fig. 4A; Supplemental Fig. S9A–F; Haesler et al. 2004; Vargha-Khadem et al. 2005; Schulz et al. 2010; Pfenning et al. 2014; Chen et al. 2016; Torres-Ruiz et al. 2019). FOXP2 is among the most widely studied human language-associated gene, with strong evidence for a role in the development and production of human spoken language and learned avian song (Enard et al. 2002; Vargha-Khadem et al. 2005; Maricic et al. 2013; Becker et al. 2015; Torres-Ruiz et al. 2019). The majority of ARs in the FOXP2-containing hotspot were located between the FOXP2 and TFEC transcription factors (Fig. 4D), a region known to contain FOXP2 regulatory elements (Becker et al. 2015; Torres-Ruiz et al. 2019). NEUROD6 is one of the most robust specialized down-regulated genes in the RA analog of all vocal learning bird lineages and the human laryngeal motor cortex (LMC) (Pfenning et al. 2014), and the ARs in this region are highly concentrated in the regulatory regions around the gene (Supplemental Fig. 9C). The ARs associated with MEF2C was particularly dense upstream of the transcription start site (Supplemental Fig. S9F). In development, MEF2C is down-regulated by FOXP2, allowing the formation of cortico-striatal connections (Chen et al. 2016). Knockdown of FOXP2 reduces cortico-striatal connectivity and ultrasonic vocalization sequence complexity in mice (Chabout et al. 2015; Castellucci et al. 2016; Chen et al. 2016). However, knockdown of both FOXP2 and MEF2C together partially rescues cortical-striatal connectivity and ultrasonic vocalization behavior (Chen et al. 2016). A second AR hotspot in the territory of MEF2C has the highest AR density, concentrated around the neurodevelopment-regulating transcription factor NR2F1 (Supplemental Fig. S9F; Armentano et al. 2007).
Figure 4.
Accelerated region distributions. (A–C) AR hotspot plots in genomes for the three vocal learning lineages using chicken as the reference for chromosome location and annotations. Each dot indicates the number of ARs in a 1-Mb window. Windows overlap and are offset from one another by 100 kb. Regions with AR density higher than random are “hotspots” and labeled in red dots. Gray vertical lines demark chromosome boundaries; chromosomes are arranged 1–28, W, Z. Genes in acceleration hotspots with known speech or neurodevelopmental functions and/or associated with convergent hotspots in vocal learners are indicated in each plot. (D) An enlarged view of the distribution of ARs per 100 kb in the FOXP2 containing acceleration hotspot. Genes in the hotspot are arrayed by strand (+ or −). FOXP2 is the only gene in this region with predicted vocalization behavior function based on coexpression (Z = 3.48). Silhouettes by Anthony Caravaggi and Ferran Sayol.
In the parrots, the highest AR density was in a region canonically disrupted in Mowat-Wilson syndrome, a developmental disorder characterized by a range of phenotypes, including speech delay (Mowat et al. 1998, 2003), and included the ZEB2 and ARHGAP15 genes (Fig. 4C; Supplemental Fig. 10L). The N2RF1 region that was the most accelerated region in oscines had the second highest AR density in parrots (Fig. 4C; Supplemental Fig. 10R). Other AR parrot hotspots encompass the transcription factors SOX6 and BCL11B (aka CTIP-2), and the transmembrane protein TENM2 (Fig. 4B; Supplemental Fig. S10I,K,Q). In the hummingbird, the same region containing TENM2 was found to have the second highest AR density (Fig. 4D); the greatest AR density was found near the NR2P and PARD3B genes. Another hummingbird AR hotspot included EFNA5 (Fig. 4C; Supplemental Fig. S11J). Hotspots in mallard include one hotspot overlapping a hummingbird hotspot on Chromosome 14 that includes RBFOX1 (Supplemental Fig. S8A).
In addition to testing for acceleration at the terminal branches of the vocal learning clades, we also tested for acceleration on the internal branches of the phylogeny where vocal learning is hypothesized to have arisen. These are the basal branches of parrots and of oscines, but we were unable to conduct a distinct test for hummingbirds because, with only one species, the branch of vocal learning origin is not distinctly captured. We also note that this data set is lacking some oscine stem lineages and so the branch of oscine origin could be further constrained by a more comprehensive sampling of genomes. We found 200 oscine branch of origin ARs and 152 parrot branch of origin ARs, and these include four AR hotspots in oscines, including a hotspot in the NR2F1-containing region observed in the clade-specific test and one hotspot in parrots (Supplemental Figs. S12, S13).
Integrating AR analysis with coexpression data to identify novel candidate genes
Our analyses above identified genes with known function in speech and vocal learning behavior, as would be expected if the analysis were recovering substantial signals of selection for vocal learning. However, we also sought to identify novel candidates for vocal learning–related selection. We therefore sought quantitative criteria to identify genes that were both significantly accelerated in vocal learning birds and shared biological features supporting a role in vocalization. We selected coexpression as the additional biological information supporting a vocalization role, as it has been hypothesized that genes that share functions tend to be expressed together (Stuart et al. 2003; Lachmann et al. 2018). We used ARCHS4, which integrates >180,000 human and mouse RNA-seq samples from the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) to predict gene function. We obtained the ARCHS4 (Lachmann et al. 2018) (accessed Nov. 20, 2019) functional predictions for all annotated protein coding genes in the chicken reference (galGal5). We searched for the possibility of strong vocal learning candidates as those genes with significant overrepresentation in ARs of one or more vocal learners (Bonferroni corrected, P < 0.05) and strong predicted vocalization behavior function by ARCHS4 coexpression (Z > 5.0) (Supplemental Table S4). This analysis identified several strong candidates, including the aforementioned known vocal learning candidates of NEUROD6 and MEF2C, as well as novel candidates NR2F1 and TENM2 (Table 1).
Table 1.
Strong candidate genes for vocal learning–related selection inferred from AR enrichment and coexpression data
If our underlying hypothesis that vocal learning birds are enriched for ARs associated with vocal learning is true, we should expect to see an excess of highly accelerated genes (118 genes) among the ARCHS4 genes with strong vocalization behavior coexpression scores (845 genes). We used the 12,894 galGal5 annotated genes with ARCHS4 score information as a background gene set. Using a hypergeometric test, we found significant enrichment (P = 0.0043), indicating that AR enriched genes are disproportionately likely to be enriched for vocalization behavior coexpression, consistent with a net boosting of confidence by combining the two criteria.
Convergent acceleration between vocal learning birds
In each vocal learning lineage, we expect that only a subset of the ARs we identify will be involved in vocal learning evolution, whereas the remainder will be involved in other adaptations. We hypothesize that convergent ARs in vocal learners, defined here as ARs that are accelerated in more than one vocal learning lineage, may prove to be greatly enriched for vocal learning related selection, because vocal learning is a shared adaptation between the lineages. Using a hypergeometric test, we found significant overrepresentation of convergent ARs across all pairwise comparisons of vocal learning bird lineages: Oscines and Parrots (observed 131, expected 50.13, P[X ≥ 131] < 1 × 10−14), Oscines and Hummingbirds (observed 44, expected 23.78, P[X ≥ 44] = 1.07 × 10−4), and Parrots and Hummingbirds (observed 50, expected 23.34, P[X ≥ 50] = 8.02 × 10−7) (Fig. 5A). One AR was shared across all three vocal learning lineages, but this did not constitute a significant enrichment over random expectation (observed 1, expected 0.90, P[X ≥ 1] = 0.59). However, this AR resides upstream of the key neurodevelopmental regulator FEZF2 (Chen et al. 2008; Rouaux and Arlotta 2010) in an intron of CADPS. Deletions encompassing the human orthologous region for this AR result in significant language deficits (de la Hoz et al. 2015; Parmeggiani et al. 2018). Additionally, both FEZF2 (rank 6th, Z = 5.19) and CADPS (rank 6th, Z = 5.22) have strong ARCHS4 predictions for vocalization behavior functionality. So, although convergence shared between all three vocal learning lineages is not significantly enriched, this specific locus is a promising candidate. Across all pairwise comparisons of waterbirds, we found four convergent ARs, one between mallard and each of the other waterbird lineages and one between core waterbirds and plovers (Supplemental Table S6). None of the waterbird convergences were significant (hypergeometric P < 0.05), but this may be due to the smaller number total ARs observed in waterbirds reducing statistical power rather than a genuine absence of convergent AR evolution in waterbirds.
Figure 5.
Vocal learning bird AR convergence. (A) Venn diagram of convergent acceleration between vocal learning bird clades. All pairwise overlaps are significantly enriched compared to random overlap (hypergeometric P < 0.05). (B) Genomic distribution of ARs convergent between two or more vocal learning bird groups. Two hotspots of acceleration (red) exceed the 95th percentile of permutation tests (horizontal line at 5 ARs/Mb) indicating significant enrichment of convergent ARs. Listed in these peaks are two candidate convergent vocal learning genes. (C) Expanded view of the highest density convergent AR hotspot, which is on the Z sex chromosome (present in both males and females). The distribution of all genes is shown in blue bars, genes with ARCHS4 Z-scores greater than 5.0 (NR2F1 and MEF2C) are highlighted on the lower gene track in red bars. Below are densities of ARs within the hotspot for each vocal learning bird lineage (single species): oscines (green), parrots (orange), and hummingbird (pink; because there is only one hummingbird AR in this region, the pink is not visible at lower resolutions). Convergent ARs are shown on a lower track in blue.
In total, 42 of 223 vocal learner convergent ARs come from two AR hotspots on Chromosomes 13 and Z (Fig. 5B; Supplemental Fig. S14A,B). The Chromosome 13 hotspot is predominantly composed of convergence between hummingbirds and parrots (eight of 10 ARs), located between TENM2 and MAT2B genes. TENM2 is a member of a gene family with important roles in motor neuron guidance (Zheng et al. 2011) and, in addition to its strong vocalization behavior coexpression discussed earlier (rank 3rd, Z = 5.56), it has been implicated in learning behavior in mice (Delprato et al. 2015). TENM2 is also associated with a cluster of great ape ARs (Kostka et al. 2018). The Chromosome Z hotspot is predominantly composed of convergent AR nucleotide sites between oscines and parrots (31 of 32 ARs) upstream of the NR2F1 transcription factor (Fig. 5C; Supplemental Fig. S14B). NR2F1 also exhibits strong vocalization behavior coexpression (rank 1st, Z = 5.83) and plays a key role in the arealization of the brain; cortical deletion of NR2F1 in mice results in major expansion of the M1 motor region to occupy most of the cortex (Armentano et al. 2007).
At the broader level of affected regions and genes, 903 genes were enriched for AR associations relative to a random background expectation in at least one of the three vocal learning lineages (hypergeometric test, P < 0.05). Eighty-two genes were enriched in two lineages and NDST3 was enriched in all three lineages (Supplemental Table S7). NDST3, which is found in a parrot AR hotspot on Chromosome 4 (Fig. 4B), is predicted to have a vocalization behavior function by ARCHS4 (Table 1). NDST3 has also been associated with schizophrenia and bipolar disorder (Zhang et al. 2016), suggesting a neurological function for the gene.
Amino acid sequence convergence has been shown to occur at higher rates in closely related species than in more distantly related species, possibly as a consequence of pleiotropy (Goldstein et al. 2015). To explore the dynamics of AR nucleotide convergence, we tested for convergence between all pairs of vocal nonlearning species. We observed significant increases in convergence of ARs over chance between closely related species relative to more distantly related species across the phylogeny (Supplemental Table S8; Supplemental Fig. S15). These results indicate that the AR distributions of closely related species are nonindependent, but more distantly related species are increasingly well modeled by independence. This phenomenon occurs in all three major avian lineages (Paleognathes, Galloanseres, and Neoaves), indicating it is common across birds and not related to vocal learning (Supplemental Fig. S15).
In light of this observation, we conducted an additional analysis to explore whether vocal learners are convergent at a higher rate than would be expected given their relatedness. We tested whether ARs from a vocal learning species (V1) were more likely to be convergent with another vocal learner (V2) or with that vocal learner's nonlearning closest relative (N); for simplicity, we denote this test as (V1 [V2, N]). For example, a test assessing whether parrot ARs are more likely to be convergent with oscine ARs or rifleman ARs is denoted (Parrot [Oscine, Rifleman]). Vocal learners shared convergent ARs at higher rates with other vocal learners than with their nearest vocal nonlearning relatives (Supplemental Fig. S16); a paired t-test (n = 5) indicates statistical support for vocal learning convergence across tests (P = 0.009). When analyzed as individual comparisons, two of the five comparisons (Parrot [Oscine, Rifleman]) (P = 0.0052) and (Hummingbird [Parrot, Rifleman]) (P = 0.017) were also statistically significant; other tests favored vocal learning convergence but with less support, (Parrot [Hummingbird, Swift]) (P = 0.34), (Oscine, [Hummingbird, Swift]) (P = 0.31), and (Hummingbird, [Oscine, Rifleman]) (P = 0.12). For the remaining tests, we lack sufficient statistical power to reject the background rate of convergence.
We then tested whether vocal learner convergence remained significant if we excluded ARs that are also shared with close nonlearning relatives of the vocal learning species. For the oscine and parrot comparison, we excluded ARs observed either in rifleman or in the manakin. For comparisons including the hummingbird, we excluded Chimney Sift ARs. Following this filtration, we found 97 Oscine-Parrot exclusive ARs (expected 41.09, filtered 34, P[X ≥ 97] = 2.89 × 10−14), 38 Oscine-Hummingbird exclusive ARs (expected 18.59, filtered 6, P[X ≥ 38] = 4.16 × 10−5), and 43 Parrot-Hummingbird exclusive ARs (expected 20.59, filtered 7, P[X ≥ 43] = 8.32 × 10−6) (Supplemental Table S6). These results are comparable in significance to the unfiltered results and continue to support greater convergence than expected under independence. Both convergent acceleration hotspots remained significant, and the convergent AR shared between all three vocal learning lineage was not filtered.
Convergent acceleration between vocal learning birds and humans
Avian vocal learning is often studied as a convergent model system for human speech. To explore this hypothesis at the level of targets of selection, we tested for AR convergence between vocal learning birds and humans. We compiled human ARs from five studies (Lindblad-Toh et al. 2005; Pollard et al. 2006; Prabhakar et al. 2006; Bird et al. 2007; Gittelman et al. 2015), for a total of 3134 human ARs ascertained from a variety of background conserved element criteria. Then, we converted the accelerated regions in human genome coordinates to the avian coordinates with liftOver (Supplemental Table S9; Kent et al. 2002) and calculated the number of avian ARs that were orthologous with human ARs. We found 64 convergent ARs between humans and at least one vocal learning bird lineage, constituting a significant enrichment for convergence between humans and each vocal learning bird lineage: oscine songbirds (observed = 23, expected = 10.15, P = 0.00031); parrots (observed = 23, expect = 11.33, P = 0.0013); and hummingbirds (observed = 18, expected = 5.52, P = 0.000017). The greatest enrichment observed in the study was between ARs convergent between two vocal learning bird lineages and humans (observed = 7, expected = 0.79, P = 1.64 × 10−5) (Fig. 6A; Supplemental Table S10). Among these 64 ARs were those near FOXP2, MEF2C, NR2F1, and EFNA5 mentioned earlier.
Figure 6.
Tests for human convergence and vocalization enrichment. (A) Venn diagram of convergence between avian and human ARs that are mappable between birds and humans. (B–E) Tests for enrichment of ARs associated with gene lists of strong relevance to human-avian convergence and vocal learning. Vocal learning bird (red) and waterbird (blue) dots indicate the observed association between ARs and genes in the list; black bars give the distribution of values expected by chance under 95% (thick) and 99.9% (thin) of cases; expected value ranges are inversely proportional to the number of ARs and to the size of the gene list. Gene lists include: (B) genes associated with one or more human accelerated regions; (C) genes identified as convergently differentially expressed between vocal learning birds’ RA song nucleus and the human laryngeal motor cortex by Pfenning et al. (2014); (D) SFARI class S genes associated with syndromic forms of autism spectrum disorders; and (E) genes identified as coexpressed with vocalization behavior GO-term genes to Z > 5.0.
Next, we tested whether there were convergent ARs in vocal learning birds and humans near the same gene locus, but not requiring the ARs to be orthologous; not all selection must be on the same genetic locus to be considered convergent. To do this, we calculated the number of vocal learning bird or waterbird ARs associated with a gene whose human ortholog is associated with a human AR. To quantify the background rate of random associations with ARs for each bird lineage, we conducted 1000 permutations, sampling conserved elements equal to the number of ARs in each acceleration test. We found significant enrichments of association between human and vocal learning bird convergent ARs (P < 0.0001, Z = 8.75), oscine songbird ARs (P < 0.0001, Z = 19.86), parrot ARs (P < 0.0001, Z = 22.57), hummingbird ARs (P < 0.0001, Z = 12.12), and the mallard ARs (P < 0.0001, Z = 5.01) (Fig. 6B), albeit a smaller enrichment than we observe with avian vocal learners. Further assessment of the terminal branches leading to vocal nonlearners revealed 15 additional bird lineages with ARs convergent with human (Supplemental Fig. S17A). However, none of these attained the fold enrichment observed in vocal learner convergent ARs, oscine ARs, or Parrot ARs with humans, and only 1 of the 22 tests (Hoatzin) exceeded the fold enrichment observed in hummingbirds (Supplemental Fig. S17A).
To test whether the differences in enrichment might be a product of the number of ARs found in each lineage, we down-sampled vocal learner ARs to the same numbers observed in waterbirds. Across 1000 replicates per vocal learner, the vocal learners were consistently enriched with ARs also specific to human relative to the waterbirds (Supplemental Fig. S18). This indicates that our results are not the product of a bias caused by the greater number of ARs found in vocal learners.
Previous studies have demonstrated convergent gene expression specializations (up- or down-regulated) in avian song and human speech brain regions (Pfenning et al. 2014). We compared the list of 52 convergent differentially expressed genes in the RA song nucleus analog of oscine songbirds, parrots, and/or hummingbird and the human LMC. We found significant enrichment (>95% of permutations) in four tests: the oscine clade, parrot clade, vocal learning convergent regions, and mallard (Fig. 6C). The oscine (P < 0.0001, Z = 4.55) and parrot (P < 0.0001, Z = 4.27) clades fell outside the range of permutation tests, but vocal learner convergent ARs (P = 0.02, Z = 2.28) and mallard ARs (P = 0.01, Z = 2.87) did not. Among nonlearner terminal branch tests, only Hoatzin and Ostrich fell outside the range of permutation tests (Supplemental Fig. S17B). The lack of enrichment of hummingbird ARs may be due to the lower ascertainment of genes differentially expressed due to cross species hybridization of the arrays in the Pfenning et al. (2014) study, the presence of only a single hummingbird in our alignment, or different selective patterns in hummingbirds relative to oscine songbirds and parrots.
Association between vocal learning bird ARs and autism spectrum disorders
ASDs are diverse in both pathology and cause, but a substantial subset of ASDs are characterized by developmental speech and language disabilities. Speech delay is one of the earliest clinical markers of ASD, and fMRI in early childhood reveals differences in speech-related brain activity between ASD probands and developmentally typical children (Redcay and Courchesne 2008). These deficits are presumed to be controlled by genetic changes in genes associated with speech development and function. Therefore, we hypothesized an enrichment of ASD genes implicated in speech in our vocal learning bird AR data sets. We compared avian genes associated with ARs to the Simons Foundation Autism Research Initiative (SFARI) category S “syndromic” in autism genes that have been shown to be associated with at least one etiology of ASD in humans, many of which involve language deficits (Mowat et al. 2003; Wang et al. 2011; Phelan and McDermid 2012; Sarasua et al. 2014). We tested both for enrichment relative to random chance by permutation of the conserved elements and found that SFARI category S genes were highly enriched for genes associated with ARs in songbirds (P < 0.0001, Z = 26.61), parrot (P < 0.0001, Z = 4.99), and the vocal learner convergent set (P < 0.0001, Z = 12.60) (Fig. 6D). There was no enrichment for SFARI class S genes in waterbirds (Fig. 6D) nor in the terminal branches of vocal nonlearners, except for the Crested Ibis and the two suboscine lineages, rifleman and two manakins (although not each manakin alone) (Supplemental Fig. S17C). The latter result is indicative of a general enrichment across the terminal branches of Passerines (both oscine and suboscine), although the fold enrichment is greater in the convergent vocal learning group than the manakin clade (Supplemental Fig. S17C).
We also tested whether the top candidate genes identified in this study supported by both strong AR enrichment and significant ARCHS4 vocalization behavior support (Table 1) are more likely to be associated with SFARI class S genes (<1% of all genes) than those predicted by ARCHS4 vocalization behavior alone. We found that two of the 16 (12.5%) best candidate genes identified in this study (Table 1) were SFARI class S genes (NR2F1 and SYT1). We further found that 23 of the 845 (2.7%) genes with an ARCHS4 vocalization behavior Z-score > 5.0 were SFARI class S genes. A Fisher's exact test to determine whether these results are from the same distribution produced a P-value = 0.07, a trend toward significance. This result suggests that the introduction of avian ARs might lead to an increase in enrichment for ASD-associated genes over ARCHS4 data alone, but the small number of best candidate genes limits our statistical power and confidence.
Association between vocal learning bird ARs and vocalization behavior–coexpressed genes
We also performed a permutation test for enrichment of ARs associated with genes that were highly coexpressed with vocalization behavior Gene Ontology (GO) term genes (Z > 5) in the ARCHS4 analysis. We found all three vocal learning lineages and their convergent grouping were strongly enriched for association with these genes in vocalization behavior (Fig. 6E). Among waterbirds, only the mallard was significantly enriched but to a lower degree than the vocal learners (Fig. 6E). Among the nonlearner terminal branches, besides the mallard, the rifleman and chimney swift also showed enrichment, but all still with lower fold enrichments than any of the vocal learners (Supplemental Fig. S17D). These findings support coexpression with vocalization-associated genes as a criterion for novel candidate discovery.
Discussion
The identification and functional interrogation of ARs has great potential for biological and therapeutic insight. Human ARs have been shown to be a valuable tool for identifying the molecular basis of key human adaptations, potentially including increased brain size and language development (Kamm et al. 2013; Oksenberg et al. 2013; Boyd et al. 2015). Our finding of elevated convergence of ARs both among vocal learning birds and between vocal learning birds and humans further support the hypothesis that vocal learning arose through convergent mechanisms. It is also the first identification of specific candidate nucleotide regulatory regions of genes with specialized brain regulation or function in vocal learning behavior. Moreover, our findings identify such regions in avian vocal learners and in humans, loci relevant to human speech and associated disorders.
A key example in the highest density AR hotspot in parrots encompasses the ZEB2 gene known to be involved in Mowat-Wilson syndrome, a neurodevelopmental disorder that causes severe speech impairment or speech absence (speech absence in >67% of patients) but has much less impact on receptive language, allowing many patients to communicate nonverbally (Ivanovski et al. 2018). Another example is FOXP2, in which a heterozygous mutation that inactivates one copy of the gene, causing an apraxia of speech (Lai et al. 2001). In addition to these genes, we identified hotspots that include other genes found in vocal learning studies, notably MEF2C and NEUROD6. FOXP2 and MEF2C play mutual regulatory and antagonistic roles in the development of vocalization systems in mammals (Chen et al. 2016). The presence of both genes in acceleration hotspots in oscine songbirds is strongly suggestive that the same pathway was a key target of vocal learning evolution in oscines as well as humans. Finding a candidate AR for NUEROD6, we could have identified the region responsible for the convergent down-regulation of NEUROD6 in the RA analog of all three avian vocal learners and the human LMC (Pfenning et al. 2014), a regulation of a transcription factor that may underlie downstream regulatory impacts. These findings are important because they allow us to link some but not all of the known vocal learning candidate genes with associated ARs that might be responsible for their specialized regulation and thus serve as key targets for future interrogation. They also serve as a valuable source of support for the viability of our method overall.
Our findings also reveal novel candidate genes and their associated ARs, including within the list of 16 of the 12,908 genes scored in ARCHS4 analysis. These novel candidates include representatives from both of the convergent acceleration hotspots (NR2F1 and TENM2) and NDST3, the only gene enriched for AR association in all three vocal learning bird groups. Perhaps NR2F1 is the most notable novel candidate gene identified. NR2F1 is a neurodevelopment regulating transcription factor and had the strongest coexpression predicted function in vocalization behavior, the highest density AR hotspot specific to vocal learning birds, and it is a SFARI class S gene for ASD (Abrahams et al. 2013). NR2F1’s role in the arealization of the brain (Armentano et al. 2007) makes it a good candidate for the expansion of forebrain size and increased pallium (i.e., cortical) neuron density in songbirds’ and parrots’ brains (Olkowicz et al. 2016), which in turn may have accommodated their extra vocal learning brain pathway (Feenders et al. 2008; Chakraborty and Jarvis 2015; Chakraborty et al. 2015).
The finding that NR2F1 and some other candidate genes are located at the edge of a gene desert is consistent with human-specific AR studies that have noted a similar pattern of enrichment of ARs in gene deserts near developmental transcription factors (Hubisz and Pollard 2014). Indeed, this is described as the “typical” human accelerated region by the originator of the method (Hubisz and Pollard 2014). It is hypothesized that these regions harbor extensive regulatory elements responsible for modulating the expression of major developmental regulators that border the gene deserts (Nobrega et al. 2003; Touceda-Suárez et al. 2020). The observed tendency for accelerated regions, both in humans and now in vocal learning birds, to occur in gene deserts lends additional support to the hypothesis that these structures may be particularly important venues for adaptive evolution.
Prior to this study, an analysis of vocal learning bird ARs was conducted on an alignment of 15 species including six vocal learning species as part of a larger comparative genomics study (Zhang et al. 2014). However, a number of technical shortcomings limited the utility of the findings of that study, including a lack of multiple test correction and lack of filtering GC-biased gene conversion, a common nonselection source of acceleration that occurs nonrandomly in the avian genome (Groenen et al. 2009; Backström et al. 2010). As a consequence of these shortcomings, any signal in the previous study was likely overwhelmed by false positive results. We controlled for all these factors. Additionally, the advances in nonreference genome alignment we used allowed us to analyze a substantially greater portion of the genome than previous studies.
Though significant progress has been made, conceptual challenges remain in extracting the most valuable and error-free possible set of ARs. A long-standing interpretive limitation of acceleration studies is the difficulty of distinguishing positive selection from loss of evolutionary constraint, as both phenomena increase the rate of substitution above the conserved rate of other taxa at the locus. For a given AR, it is difficult to exclude the possibility that the observed pattern of acceleration is the result of loss of constraint. For shorter divergence time spans (∼<10 million years), a test of whether the rate of acceleration exceeds the neutral rate is often applied (Nei and Gojobori 1986; Pollard et al. 2010). However, such a test assumes a single uniform selective regime, which may be inappropriate over the time spans being assessed in this study (30–50 million years), where an adaptive event might lead the rate of molecular evolution to shift from conservation to acceleration and back to conservation. Further, loss of function would be consistent with some of the findings on gene expression, where vocal learning bird RA and human LMC have loss of expression of NEUROD6 and other neural development genes.
Another conceptual challenge we encountered was our finding that the number of ARs in a lineage was positively correlated with branch length and the number of species in the clade being assessed. The branch length finding is consistent with a relatively stable rate of positive selection events within each lineage. It is broadly consistent with the finding that most human ARs are shared with Neanderthals (Hubisz and Pollard 2014). The effect of the number of species in the clade is more complex where, in from one to four species, we see an increase in the correct identification of regions as accelerated, and an increase in power beyond four species. There also appears to be a shift toward ascertaining selection events shared across the clade, meaning they either took place early in the clade's evolution or were convergently evolved in multiple lineages within the clade. For this study, the skew in AR ascertainment toward the base of the clade is desirable because we are interested in a trait shared across all members of the clade.
Looking forward, technical advances will almost certainly be a major source of improvement in acceleration studies. This includes higher throughput sequencing of many more species per lineage and improvements in genome sequencing and assembly allowing for more complete and error-free assemblies (Rhie et al. 2021). Likewise, the proliferation of tissue-specific epigenomic methods for identifying functional elements, such as ChIP-seq and ATAC-seq, present great potential for further targeting the most relevant ARs in this study.
The ascertainment of convergent ARs has been a topic of great interest in the recent literature because of their potential to reveal selection underlying trait evolution (Hu et al. 2019; Kowalczyk et al. 2019; Partha et al. 2019; Sackton et al. 2019). These have great potential to further expand this work and potentially reveal greater patterns of convergence through improved statistical power. Although there have not yet been rigorous comparisons of these methods to ascertain which are most useful and reliable, the new methods represent an important potential area for growth in the field. One recent method proposes a novel Bayesian approach to identify both convergent and nonconvergent ARs from a convergently evolving trait group in a single analysis (Hu et al. 2019). However, this implementation of phyloP would not be appropriate for our analyses as it measures the unweighted average of acceleration across all of the convergent lineages (Pollard et al. 2006) and could recover acceleration in the lineage with the most mutations on average (Hu et al. 2019). To avoid such a bias, we ran phyloP independently on each convergent lineage and then intersected the results to identify convergent ARs.
A further consideration for studies that attempt to compare between AR identification methods should consider our observation that closely related lineages are more likely to be convergently accelerated than more distantly related lineages. Greater amino acid convergence (Goldstein et al. 2015) and AR convergence (this study) presents an interpretive challenge: true positive convergence events occurring at a rate greater than chance might still be, in some sense, expected.
Although convergent acceleration between all pairs of vocal learners occurred at higher than expected rates, we did not find any ARs shared between all three vocal learning bird lineages and humans. This is consistent with other notable examples of convergence at the DNA level, such as pigmentation regulation by the melanocortin 1 receptor (MC1R). MC1R variants are responsible for pale pigmentation in a range of species, including birds (Mundy et al. 2004), lizards (Rosenblum et al. 2004), white “spirit bear” American black bears (Ursus americanus) (Ritland et al. 2001), and Florida Gulf Coast beach mice (Peromyscus polionotus) (Hoekstra et al. 2006). However, similarly light-coated Florida Atlantic Coast beach mice (Peromyscus polionotus) and polar bears (Ursus maritimus) both lack derived MC1R variants and are thought to have evolved their light coats through non-MC1R-related mechanisms (Hoekstra et al. 2006; Miller et al. 2012). So, whereas molecular convergence on the same gene and sometimes same nucleotide sites does occur, it is not universal. This nonuniversality highlights the value of comparisons extending across many species and analyses that look at both types of convergences. By including many lineages in our analyses, we can identify more candidate component genes underlying a complex trait.
Another consideration for all studies on evolution of avian vocal learning is that, given the tree topology (Fig. 1B), whether vocal learning evolved three times in birds, as assumed in this study, or twice—once in hummingbirds and once in the ancestor of parrots and songbirds—and then subsequently was lost twice in suboscines (once in the rifleman clade and once in the larger clade including manakins) (Jarvis et al. 2014). Whereas both hypotheses have merits, the three-origins hypothesis is more parsimonious, as the branch ancestral to parrots and all songbirds (oscine and suboscine) is very short, allowing only a very narrow time span where vocal learning could have evolved substantially, and the parrot vocal learning brain pathway has two parallel vocal learning systems whereas songbirds and hummingbirds have just one, indicating that there had to be at least further independent evolution in parrots (Chakraborty and Jarvis 2015; Chakraborty et al. 2015). Nevertheless, both three- or two-origin hypotheses are consistent with the convergent ARs in parrots and songbirds with hummingbirds and humans; and if two losses occurred in rifleman and suboscines, this is still consistent with the presence of ARs in songbirds and parrots associated with the presence of vocal learning. Our findings are also consistent with the continuum hypothesis of vocal learning (Arriaga et al. 2012; Petkov and Jarvis 2012; Jarvis 2019), where the advanced vocal learners have more advanced ARs not found in more limited vocal learners. Future investigations with more genomes and phenotypic vocalization characterization across species would inform whether there are more continuous AR changes across species with a continuous vocal learning trait.
In conclusion, this study provides an extensive collection of positively selected loci and specific nucleotide changes in avian and human vocal learning lineages to be mined and assessed by future studies. The discovery of acceleration hotspots, encompassing both widely recognized genes such as FOXP2 and functionally plausible novel and convergent candidates such as NR2F1 and NEUROD6 in vocal learning behavior, point to new evolutionarily informed avenues for experimental testing. Our finding that some AR hotspots in vocal learning birds are in the same genomic regions implicated in human speech pathologies, including Mowat-Wilson disorder and verbal ASD, also serve of candidate genomic regions to study such speech deficits.
Methods
Sample selection
We selected 33 published avian genomes generated using short reads or Sanger sequencing for this analysis (Supplemental Table S1), except for zebra finch (Korlach et al. 2017), Anna's hummingbird (Korlach et al. 2017), and chicken v5 (International Chicken Genome Sequencing Consortium 2004), which used long reads and thereby had fewer gaps and were more complete. We prioritized the inclusion of species in vocal learning clades (oscine songbirds, parrots, and hummingbirds) and their near vocal nonlearning relatives (suboscine songbirds, nonhummingbird Caprimulgiformes, and falcons) (Jarvis et al. 2014). We selected the remaining samples to provide a diverse representation of the major avian clades prioritizing published genomes with greater assembly contiguity (Supplemental Table S1) (Romanov et al. 2011; Cai et al. 2013; Seabury et al. 2013; Shapiro et al. 2013; Doyle et al. 2014; Jarvis et al. 2014; Zhang et al. 2014; Koepfli et al. 2015).
Whole-genome alignment
To reduce the risk of false alignments and reduce the computational work required for genome alignment, we soft-masked repeats in the genomes with RepeatMasker (Smit et al. 2013). We aligned the genomes using a reference-free approach, Cactus (https://github.com/ComparativeGenomicsToolkit/cactus/), commit 95f1c43c9740201aec52844c085cc3bb92fb5757 (Paten et al. 2011), on the AWS cloud platform. Progressive Cactus alignments require a guide tree to establish the relationships between taxa. To generate this guide tree, we used the phylogeny of Jarvis et al. (2014) as a backbone and added species not present in that alignment on the basis of their location in the Prum et al. (2015) phylogeny with estimated branch lengths. Although a guide tree is required for Progressive Cactus alignments, minor deviations from realistic phylogenies do not result in substantially different alignments (Armstrong et al. 2020).
Conserved element identification
We generated a neutral model of sequence evolution using a predefined tree topology and estimated branch lengths and substitution rates from fourfold degenerate sites using PHAST phyloFit (version 1.4) (Supplemental File S1; Siepel et al. 2005; Hubisz et al. 2011). We identified fourfold degenerate sites using the chicken reference (galGal5) genome annotation (International Chicken Genome Sequencing Consortium 2004). Previous studies have shown anomalous tree topologies inferred from coding sequences in birds, most likely as a result of nonhomogeneous rates of GC-biased gene conversion across the avian genome (Jarvis et al. 2014; Reddy et al. 2017). However, our use of a predefined topology inferred from more typical loci should minimize the risk of error due to misinterpreting sequence homoplasy as sequence homology.
Having established a null hypothesis for neutral evolution in this data set, we identified conserved elements in the 21 vocal nonlearning birds using PHAST phastCons (version 1.4) (Siepel et al. 2005; Hubisz et al. 2011). We set the –target-coverage flag to 0.25, adjusted from Hubisz et al. (2011) to account for the smaller genome size of birds relative to mammals and the –expected-length flag to 20 (Siepel et al. 2005; Hubisz et al. 2011). We excluded: conserved elements <100 bp in keeping with previous AR studies (Pollard et al. 2006; Hubisz et al. 2011; Hubisz and Pollard 2014); conserved elements with fewer than 15 vocal nonlearning species; and vocal learning tests with less than either five oscines, three parrots, or one hummingbird.
In a biological control analysis, we also tested for acceleration in four independent gains in waterbirds (mallard; crested ibis and emperor penguin; American flamingo; killdeer). We followed the same methods as used for vocal learners, identifying conserved elements from all 28 nonwaterbird species with phastCons (version 1.4) (Siepel et al. 2005; Hubisz et al. 2011). The only difference from the vocal learner grouping is that we increased the minimum number of nontrait species of interest (nonwaterbird) individuals present in the alignment at a given region from 15 to 20. We did this because there were more nonwaterbird species (28) than vocal nonlearner (21) species. Thus, we required a similar fraction of species without the trait of interest to be included in the alignment rather than the same absolute number of species.
Filtering paralogous alignments
To ensure that our analysis used 1-to-1 orthologous sequences, we masked regions of the alignment where any of the species being tested for acceleration included multiple sequences aligned to the same position in the chicken genome. Thus, for all vocal learning species, except the zebra finch, we masked any region with two or more aligned sequences. The zebra finch assembly we used (Tgut_diploid_1.0) includes a large fraction of the genome where the maternal and paternal haplotypes are both included as separate scaffolds (Korlach et al. 2017). In those regions, the zebra finch assembly is expected to have two sequences that are true 1-to-1 orthologs of the chicken assembly. To accommodate this idiosyncrasy of the zebra finch assembly, we masked sites where the zebra finch assembly had three or more aligned sequences.
In addition to cases where both an ortholog and a paralog are present in the vocal learner's assembly, it is possible that only a paralog of the chicken sequence may be present in a vocal learning species. To detect and filter this second type of potential paralogous alignment, we developed a genome-wide scan for alignment blocks composed of two highly divergent groups of sequences. In a typical orthologous alignment, the pairwise differences between samples had a unimodal distribution. However, in paralogous alignments we expect a bimodal distribution with one mode for pairwise differences between orthologous sequences and a second mode for pairwise differences between paralogous sequences (Supplemental Fig. S19). To identify such alignments, we calculated the pairwise sequence divergence between all samples in 1-kb windows across the alignment. To distinguish between unimodal and bimodal distributions, we sorted the distribution of pairwise differences between the samples and identified the maximum difference between sequential comparisons. We restricted our analysis to the 15th to 85th percentile of the pairwise differences to prevent a single outlier sample from producing a false paralog identification. We define strong evidence of a bimodal distribution as cases where the largest difference between sequential pairwise differences was more than 50% of the median pairwise difference and exclude those regions from the analysis.
GC-biased gene conversion
GC-biased gene conversion (gBGC) is a recombination-driven mutation process that can lead to locally high substitution rates in a lineage without positive selection. These sites represent a major source of ARs that are not under positive selection (Katzman et al. 2010; Capra et al. 2013). We used PHAST phastBias (version 1.4) (Hubisz et al. 2011; Capra et al. 2013) to identify gBGC events in each branch of the phylogeny. In keeping with the recommendation of the authors of the program, we consider any region with a score >0.5 to be impacted by gBGC (Hubisz et al. 2011). In our tests for positive selection, we exclude any region identified as impacted by gBGC on a branch being tested for acceleration.
Accelerated region identification
We used PHAST phyloP -ACC with the likelihood ratio test method (version 1.4) (Pollard et al. 2006; Hubisz et al. 2011) to test for acceleration in each of the three vocal learning clades. Each analysis was conducted with a single clade of vocal learning birds as the foreground group and the nonlearning birds as the background group. The other two vocal learning groups were excluded from the analysis to avoid biasing against detecting convergent acceleration specific to a vocal learning clade. We conducted multiple test corrections using a Benjamini–Hochberg resampling with RPHAST (Hubisz et al. 2011). We generated 100,000 100-bp resamplings per genome from the nonlearner conserved regions. Each individual resampling was conducted within a single chromosome (using chicken chromosome designation) due to computational limitations; the number of resamplings per chromosome was proportional to the size of the chromosome, so larger chromosomes were sampled more frequently. We considered significant acceleration to be regions with a Benjamini–Hochberg false discovery rate ≤0.05.
AR hotspot density analysis
We scanned the distribution of conserved elements and ARs across the genome in 1-Mb overlapping windows separated by a 100-kb step. To test whether ARs and conserved elements were correlated, we generated a scatterplot of conserved elements versus ARs for each window and calculated the linear regression R2 (Supplemental Fig. S7). To identify outlier regions of AR density, that is, “acceleration hotspots,” we employed a strict uniform cutoff inferred through permutation testing for each AR analysis (oscine, parrot, hummingbird, and vocal learning convergent) (Figs. 4A–C, 5B). For each AR analysis, we conducted 100 permutation tests: we randomly assigned N conserved elements to be accelerated, where N is the number of ARs observed in the data. Then, we scanned the genome with a -Mb sliding window in 100-kb steps to determine the genome-wide maximum AR density per Mb. If a region had an AR density >95% of the genome-wide maxima observed in the permutation tests, we considered it to be in an acceleration hotspot.
Associating ARs with genes
To assess the potential biological impacts of ARs and test for enrichments of certain categories of traits among the ARs, we predicted the genes most likely to be impacted by each AR. To achieve this, we first filtered the chicken reference genome annotation (GCF_000002315.4) (International Chicken Genome Sequencing Consortium 2004) to include genes labeled as protein-, tRNA-, and rRNA-coding to mitigate the potential impact of incorrect annotations. Then, we classified ARs based on their proximity to the annotated genes: (1) First, we identified all coding ARs, defined here as ARs that at least partly overlap an exon, and considered them associated with the gene coded for; (2) we classified the remaining ARs as noncoding, and applied GREAT's “basal plus extension” criteria (McLean et al. 2010) using custom scripts. This criterion predicts a regulatory region for each gene. First, for each gene we define a basal region from 5 kb upstream of to 1 kb downstream from each gene's transcription start site that is automatically associated with that gene regardless of the proximity of other genes. Then, we extended the predicted regulatory region to the nearest upstream and downstream basal regions or to a maximum distance of 1 Mb in each direction. Each noncoding AR was assigned to all genes whose regulatory regions it overlapped. In most cases, this means a noncoding AR is associated with two genes, one upstream and one downstream, but ARs in a gene's basal region (defined above) or in very low gene density regions of the genome may be associated with one or zero genes (Supplemental Fig. S20).
Candidate gene identification using predicted ontology
We used ARCHS4 (Lachmann et al. 2018) to identify genes that had gene coexpression patterns consistent with different behavior functions. ARCHS4 compiles publicly available human and mouse RNA-seq data from a wide range of tissue types and uses this data to identify coexpression between genes. Because greater coexpression can be indicative of shared function (Stuart et al. 2003), this coexpression information is leveraged to predict novel functions of genes by identifying the Gene Ontology terms with coexpression patterns similar to the gene of interest (Lachmann et al. 2018).
First, we downloaded ARCHS4 predicted GO term data for all genes in the ARCHS4 database (12,963 genes) and identified genes with vocalization behavior coexpression Z-scores > 5. Second, we calculated per gene AR overrepresentation for each acceleration test by conducting a hypergeometric test for enrichment of AR association with the gene relative to the number of conserved elements associated with the gene. We considered a gene strongly enriched if it had a Bonferroni-corrected P < 0.05. We took the intersection of these two criteria, ARCHS4 Vocalization Behavior Z > 5 and AR association enrichment P < 0.05 corrected, to be a strong candidate gene list.
Enrichment of ARs in biologically relevant gene lists
To test for enrichment of ARs associated with a list of genes (e.g., SFARI class S genes [Abrahams et al. 2013]) we first compared the number of observed associations with genes in the gene list across all ARs in a lineage (see Methods, “Associating ARs with genes”). Then, we conducted 1000 permutation tests, each of which randomly sample N post-quality filtering conserved elements (see Methods, “Conserved element identification,” “Filtering paralogous alignments,” “GC-biased gene Conversion”), where N is the number of observed ARs across the genome. Then, we calculated the number of associations with the gene list in each permutation test and used that data to estimate expected rates of association. The expected value given in Figure 6 is the median value of the permutation tests. Fold enrichments are given relative to the median of the permutation test to allow joint visualization across tests with differing numbers of ARs and hence differing expected values. Single tailed P-values were calculated as the fraction of permutations with fewer AR associations than the observed value in the real data.
Increasing numbers of ARs in the observed data tend to result in proportionally smaller differences between permutation tests and hence have greater statistical power, which is expected. However, we also wanted to test whether having more ARs might introduce a bias that would lead to more AR-rich vocal learners being more enriched for AR associations than control groups. To test this hypothesis, we randomly sampled without replacement from the AR-rich vocal learning lineages until we had ascertained a number of ARs equal to the total number ARs found in the AR-poor waterbird lineages. Then, we calculated the number of gene associations for the down-sampling. We conducted 1000 resamplings per vocal learning lineage and compared the results to the waterbird observed and expected values (Supplemental Fig. S18).
Testing for enrichment in the rate of convergence between lineages
We conducted two tests for enrichment of convergent acceleration between lineages, one which assumes independence and a second that attempts to ascertain whether convergence exceeds the degree expected given the phylogenetic relationship between the phenotypically convergent species. When assuming independence, we filtered to only include conserved elements that passed all quality criteria (described above) for the lineage being compared and conducted hypergeometric tests for enrichment of convergent ARs on the remaining loci using the SciPy package in Python version 2.7.3.
However, because we observed that closely related species frequently shared convergent ARs at a greater rate than would be expected under an assumption of independence, we conducted an additional more stringent three-lineage test. We tested for convergence between an outgroup vocal learner and an ingroup vocal learner, relative to the convergence between the outgroup vocal learner and an ingroup nonlearner. To insure uniform sampling, we first filtered to include the sites that pass filtering criteria for all three lineages being tested. Then, we calculate the rate of convergence between the outgroup and each of the ingroups. Finally, we applied a Fisher's exact test to determine whether the rate of convergence between the outgroup and each of the ingroups was significantly different.
Software availability
All scripts used for data processing and analysis are available at GitHub (https://github.com/jacahill/AR_Tools) and as Supplemental Code.
Supplementary Material
Acknowledgments
Funding was provided by the Howard Hughes Medical Institute and by Rockefeller University start-up funds to E.D.J., and the National Institutes of Health (1U01HL137183-01 and 5U54HG007990) to B.P. and D.H. We thank Mark Diekhans for facilitating the hosting of the genome alignment.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275989.121.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. 2013. SFARI gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism 4: 36. 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armentano M, Chou S-J, Tomassy GS, Leingärtner A, O'Leary DDM, Studer M. 2007. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci 10: 1277–1286. 10.1038/nn1958 [DOI] [PubMed] [Google Scholar]
- Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, Fang Q, Xie D, Feng S, Stiller J, et al. 2020. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587: 246–251. 10.1038/s41586-020-2871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriaga G, Zhou EP, Jarvis ED. 2012. Of mice, birds, and men: the mouse ultrasonic song system has some features similar to humans and song-learning birds. PLoS One 7: e46610. 10.1371/journal.pone.0046610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Ost T, Schneider M, Kempenaers B, et al. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res 20: 485–495. 10.1101/gr.101410.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Devanna P, Fisher SE, Vernes SC. 2015. A chromosomal rearrangement in a child with severe speech and language disorder separates FOXP2 from a functional enhancer. Mol Cytogenet 8: 69. 10.1186/s13039-015-0173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CP, Stranger BE, Liu M, Thomas DJ, Ingle CE, Beazley C, Miller W, Hurles ME, Dermitzakis ET. 2007. Fast-evolving noncoding sequences in the human genome. Genome Biol 8: R118. 10.1186/gb-2007-8-6-r118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis JJ, Okanoya K, Scharff C. 2010. Twitter evolution: converging mechanisms in birdsong and human speech. Nat Rev Neurosci 11: 747–759. 10.1038/nrn2931 [DOI] [PubMed] [Google Scholar]
- Booker BM, Friedrich T, Mason MK, VanderMeer JE, Zhao J, Eckalbar WL, Logan M, Illing N, Pollard KS, Ahituv N. 2016. Bat accelerated regions identify a bat forelimb specific enhancer in the HoxD locus. PLoS Genet 12: e1005738. 10.1371/journal.pgen.1005738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, Wray GA, Silver DL. 2015. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol 25: 772–779. 10.1016/j.cub.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qian X, Lang Y, Luo Y, Xu J, Pan S, Hui Y, Gou C, Cai Y, Hao M, et al. 2013. Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude. Genome Biol 14: R29. 10.1186/gb-2013-14-3-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Hubisz MJ, Kostka D, Pollard KS, Siepel A. 2013. A model-based analysis of GC-biased gene conversion in the human and chimpanzee genomes. PLoS Genet 9: e1003684. 10.1371/journal.pgen.1003684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci GA, McGinley MJ, McCormick DA. 2016. Knockout of Foxp2 disrupts vocal development in mice. Sci Rep 6: 23305. 10.1038/srep23305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabout J, Sarkar A, Dunson DB, Jarvis ED. 2015. Male mice song syntax depends on social contexts and influences female preferences. Front Behav Neurosci 9: 76. 10.3389/fnbeh.2015.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Jarvis ED. 2015. Brain evolution by brain pathway duplication. Philos Trans R Soc Lond B Biol Sci 370: 20150056. 10.1098/rstb.2015.0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Walløe S, Nedergaard S, Fridel EE, Dabelsteen T, Pakkenberg B, Bertelsen MF, Dorrestein GM, Brauth SE, Durand SE, et al. 2015. Core and shell song systems unique to the parrot brain. PLoS One 10: e0118496. 10.1371/journal.pone.0118496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. 2008. The Fezf2–Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci 105: 11382–11387. 10.1073/pnas.0804918105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Kuo H-Y, Bornschein U, Takahashi H, Chen S-Y, Lu K-M, Yang H-Y, Chen G-M, Lin J-R, Lee Y-H, et al. 2016. Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat Neurosci 19: 1513–1522. 10.1038/nn.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson G, Rogers S, Munson J, Smith M, Winter J, Greenson J, Donaldson A, Varley J. 2010. Randomized, controlled trial of an intervention for toddlers with autism: the Early Start Denver Model. Pediatrics 125: e17–e23. 10.1542/peds.2009-0958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Hoz AB, Maortua H, García-Rives A, Martínez-González MJ, Ezquerra M, Tejada M-I. 2015. 3p14 de novo interstitial microdeletion in a patient with intellectual disability and autistic features with language impairment: a comparison with similar cases. Case Rep Genet 2015: 876348. 10.1155/2015/876348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprato A, Bonheur B, Algéo MP, Rosay P, Lu L, Williams RW, Crusio WE. 2015. Systems genetic analysis of hippocampal neuroanatomy and spatial learning in mice. Genes Brain Behav 14: 591–606. 10.1111/gbb.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. 1999. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci 22: 567–631. 10.1146/annurev.neuro.22.1.567 [DOI] [PubMed] [Google Scholar]
- Doyle JM, Katzner TE, Bloom PH, Ji Y, Wijayawardena BK, DeWoody JA. 2014. The genome sequence of a widespread apex predator, the golden eagle (Aquila chrysaetos). PLoS One 9: e95599. 10.1371/journal.pone.0095599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CSL, Wiebe V, Kitano T, Monaco AP, Pääbo S. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869–872. 10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. 2008. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS One 3: e1768. 10.1371/journal.pone.0001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittelman RM, Hun E, Ay F, Madeoy J, Pennacchio L, Noble WS, Hawkins DR, Akey JM. 2015. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res 25: 1245–1255. 10.1101/gr.192591.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein RA, Pollard ST, Shah SD, Pollock DD. 2015. Nonadaptive amino acid convergence rates decrease over time. Mol Biol Evol 32: 1373–1381. 10.1093/molbev/msv041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MAM, Wahlberg P, Foglio M, Cheng HH, Megens H-J, Crooijmans RPMA, Besnier F, Lathrop M, Muir WM, Wong GK-S, et al. 2009. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res 19: 510–519. 10.1101/gr.086538.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C. 2004. FoxP2 expression in avian vocal learners and non-learners. J Neurosci 24: 3164–3175. 10.1523/JNEUROSCI.4369-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Rivas MV, Ward JM, Okanoya K, Jarvis ED. 2012. Convergent differential regulation of parvalbumin in the brains of vocal learners. PLoS One 7: e29457. 10.1371/journal.pone.0029457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313: 101–104. 10.1126/science.1126121 [DOI] [PubMed] [Google Scholar]
- Hu Z, Sackton TB, Edwards SV, Liu JS. 2019. Bayesian detection of convergent rate changes of conserved noncoding elements on phylogenetic trees. Mol Biol Evol 36: 1086–1100. 10.1093/molbev/msz049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS. 2014. Exploring the genesis and functions of Human Accelerated Regions sheds light on their role in human evolution. Curr Opin Genet Dev 29: 15–21. 10.1016/j.gde.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinformatics 12: 41–51. 10.1093/bib/bbq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Ivanovski I, Djuric O, Caraffi SG, Santodirocco D, Pollazzon M, Rosato S, Cordelli DM, Abdalla E, Accorsi P, Adam MP, et al. 2018. Phenotype and genotype of 87 patients with Mowat–Wilson syndrome and recommendations for care. Genet Med 20: 965–975. 10.1038/gim.2017.221 [DOI] [PubMed] [Google Scholar]
- Jarvis ED. 2019. Evolution of vocal learning and spoken language. Science 366: 50–54. 10.1126/science.aax0287 [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SYW, Faircloth BC, Nabholz B, Howard JT, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346: 1320–1331. 10.1126/science.1253451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm GB, Pisciottano F, Kliger R, Franchini LF. 2013. The developmental brain gene NPAS3 contains the largest number of accelerated regulatory sequences in the human genome. Mol Biol Evol 30: 1088–1102. 10.1093/molbev/mst023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman S, Kern AD, Pollard KS, Salama SR, Haussler D. 2010. GC-biased evolution near human accelerated regions. PLoS Genet 6: e1000960. 10.1371/journal.pgen.1000960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, et al. 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep 10: 112–122. 10.1016/j.celrep.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli K-P, Paten B, Genome 10K Community of Scientists, O'Brien SJ. 2015. The Genome 10K Project: a way forward. Annu Rev Anim Biosci 3: 57–111. 10.1146/annurev-animal-090414-014900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin C-S, Howard JT, Audet J-N, Cantin L, Jarvis ED. 2017. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience 6: 1–16. 10.1093/gigascience/gix085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka D, Holloway AK, Pollard KS. 2018. Developmental loci harbor clusters of accelerated regions that evolved independently in ape lineages. Mol Biol Evol 35: 2034–2045. 10.1093/molbev/msy109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A, Meyer WK, Partha R, Mao W, Clark NL, Chikina M. 2019. RERconverge: an R package for associating evolutionary rates with convergent traits. Bioinformatics 35: 4815–4817. 10.1093/bioinformatics/btz468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, Silverstein MC, Ma'ayan A. 2018. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun 9: 1366. 10.1038/s41467-018-03751-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. 2001. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413: 519–523. 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- Law J, Boyle J, Harris F, Harkness A, Nye C. 2000. Prevalence and natural history of primary speech and language delay: findings from a systematic review of the literature. Int J Lang Comm Dis 35: 165–188. 10.1080/136828200247133 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438: 803–819. 10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Liu W, Wada K, Jarvis ED, Nottebohm F. 2013. Rudimentary substrates for vocal learning in a suboscine. Nat Commun 4: 2082. 10.1038/ncomms3082 [DOI] [PubMed] [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157: 785–794. 10.1016/j.cell.2014.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Huizinga NA, Friedrich SR, Wirthlin M, Mello CV. 2018. The constitutive differential transcriptome of a brain circuit for vocal learning. BMC Genomics 19: 231. 10.1186/s12864-018-4578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T, Günther V, Georgiev O, Gehre S, Ćurlin M, Schreiweis C, Naumann R, Burbano HA, Meyer M, Lalueza-Fox C, et al. 2013. A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Mol Biol Evol 30: 844–852. 10.1093/molbev/mss271 [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, et al. 2012. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci 109: E2382–E2390. 10.1073/pnas.1210506109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat DR, Croaker GD, Cass DT, Kerr BA, Chaitow J, Adès LC, Chia NL, Wilson MJ. 1998. Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J Med Genet 35: 617–623. 10.1136/jmg.35.8.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat DR, Wilson MJ, Goossens M. 2003. Mowat-Wilson syndrome. J Med Genet 40: 305–310. 10.1136/jmg.40.5.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy NI, Badcock NS, Hart T, Scribner K, Janssen K, Nadeau NJ. 2004. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science 303: 1870–1873. 10.1126/science.1093834 [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. 10.1093/oxfordjournals.molbev.a040410 [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. 10.1126/science.1088328 [DOI] [PubMed] [Google Scholar]
- Oksenberg N, Stevison L, Wall JD, Ahituv N. 2013. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet 9: e1003221. 10.1371/journal.pgen.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkowicz S, Kocourek M, Lučan RK, Porteš M, Fitch WT, Herculano-Houzel S, Němec P. 2016. Birds have primate-like numbers of neurons in the forebrain. Proc Natl Acad Sci 113: 7255–7260. 10.1073/pnas.1517131113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani G, Buldrini B, Fini S, Ferlini A, Bigoni S. 2018. A new 3p14.2 microdeletion in a patient with intellectual disability and language impairment: case report and review of the literature. Mol Syndromol 9: 175–181. 10.1159/000489842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partha R, Kowalczyk A, Clark NL, Chikina M. 2019. Robust method for detecting convergent shifts in evolutionary rates. Mol Biol Evol 36: 1817–1830. 10.1093/molbev/msz107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten B, Earl D, Nguyen N, Diekhans M, Zerbino D, Haussler D. 2011. Cactus: algorithms for genome multiple sequence alignment. Genome Res 21: 1512–1528. 10.1101/gr.123356.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov CI, Jarvis ED. 2012. Birds, primates, and spoken language origins: behavioral phenotypes and neurobiological substrates. Front Evol Neurosci 4: 12. 10.3389/fnevo.2012.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G, et al. 2014. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346: 1256846. 10.1126/science.1256846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE. 2012. The 22q13.3 deletion syndrome (Phelan-McDermid syndrome). Mol Syndromol 2: 186–201. 10.1159/000334260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, Kern AD, Dreszer T, Katzman S, Siepel A, Pedersen JS, Bejerano G, Baertsch R, et al. 2006. Forces shaping the fastest evolving regions in the human genome. PLoS Genet 2: e168. 10.1371/journal.pgen.0020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. 2006. Accelerated evolution of conserved noncoding sequences in humans. Science 314: 786. 10.1126/science.1130738 [DOI] [PubMed] [Google Scholar]
- Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, Lemmon AR. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526: 569–573. 10.1038/nature15697 [DOI] [PubMed] [Google Scholar]
- Redcay E, Courchesne E. 2008. Deviant functional magnetic resonance imaging patterns of brain activity to speech in 2–3-year-old children with autism spectrum disorder. Biol Psychiatry 64: 589–598. 10.1016/j.biopsych.2008.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S, Kimball RT, Pandey A, Hosner PA, Braun MJ, Hackett SJ, Han K-L, Harshman J, Huddleston CJ, Kingston S, et al. 2017. Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst Biol 66: 857–879. 10.1093/sysbio/syx041 [DOI] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Gedman GL, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland K, Newton C, Marshall HD. 2001. Inheritance and population structure of the white-phased “Kermode” black bear. Curr Biol 11: 1468–1472. 10.1016/S0960-9822(01)00448-1 [DOI] [PubMed] [Google Scholar]
- Romanov MN, Dodgson JB, Gonser RA, Tuttle EM. 2011. Comparative BAC-based mapping in the white-throated sparrow, a novel behavioral genomics model, using interspecies overgo hybridization. BMC Res Notes 4: 211. 10.1186/1756-0500-4-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum EB, Hoekstra HE, Nachman MW. 2004. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution (N Y) 58: 1794–1808. 10.1111/j.0014-3820.2004.tb00462.x [DOI] [PubMed] [Google Scholar]
- Rouaux C, Arlotta P. 2010. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nat Neurosci 13: 1345–1347. 10.1038/nn.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Grayson P, Cloutier A, Hu Z, Liu JS, Wheeler NE, Gardner PP, Clarke JA, Baker AJ, Clamp M, et al. 2019. Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364: 74–78. 10.1126/science.aat7244 [DOI] [PubMed] [Google Scholar]
- Sarasua SM, Dwivedi A, Boccuto L, Chen C-F, Sharp JL, Rollins JD, Collins JS, Rogers RC, Phelan K, DuPont BR. 2014. 22q13.2q13.32 genomic regions associated with severity of speech delay, developmental delay, and physical features in Phelan-McDermid syndrome. Genet Med 16: 318–328. 10.1038/gim.2013.144 [DOI] [PubMed] [Google Scholar]
- Schulz SB, Haesler S, Scharff C, Rochefort C. 2010. Knockdown of FoxP2 alters spine density in Area X of the zebra finch. Genes Brain Behav 9: 732–740. 10.1111/j.1601-183X.2010.00607.x [DOI] [PubMed] [Google Scholar]
- Seabury CM, Dowd SE, Seabury PM, Raudsepp T, Brightsmith DJ, Liboriussen P, Halley Y, Fisher CA, Owens E, Viswanathan G, et al. 2013. A multi-platform draft de novo genome assembly and comparative analysis for the Scarlet Macaw (Ara macao). PLoS One 8: e62415. 10.1371/journal.pone.0062415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Kronenberg Z, Li C, Domyan ET, Pan H, Campbell M, Tan H, Huff CD, Hu H, Vickrey AI, et al. 2013. Genomic diversity and evolution of the head crest in the rock pigeon. Science 339: 1063–1067. 10.1126/science.1230422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley R, Green P. 2013. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Stuart JM, Segal E, Koller D, Kim SK. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255. 10.1126/science.1087447 [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz R, Benítez-Burraco A, Martínez-Lage M, Rodríguez-Perales S, García-Bellido P. 2019. Functional characterization of two enhancers located downstream FOXP2. BMC Med Genet 20: 65. 10.1186/s12881-019-0810-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touceda-Suárez M, Kita EM, Acemel RD, Firbas PN, Magri MS, Naranjo S, Tena JJ, Gómez-Skarmeta JL, Maeso I, Irimia M. 2020. Ancient genomic regulatory blocks are a source for regulatory gene deserts in vertebrates after whole genome duplications. Mol Biol Evol 37: 2857–2864. 10.1093/molbev/msaa123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargha-Khadem F, Gadian DG, Copp A, Mishkin M. 2005. FOXP2 and the neuroanatomy of speech and language. Nat Rev Neurosci 6: 131–138. 10.1038/nrn1605 [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, et al. 2011. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet 20: 3093–3108. 10.1093/hmg/ddr212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM, Li D, Eagleton C. 1997. Projections of the dorsomedial nucleus of the intercollicular complex (DM) in relation to respiratory-vocal nuclei in the brainstem of pigeon (Columba livia) and zebra finch (Taeniopygia guttata). J Comp Neurol 377: 392–413. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339: 456–460. 10.1126/science.1230835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. 10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lu W, Wang Z, Ni J, Zhang J, Tang W, Fang Y. 2016. A comprehensive analysis of NDST3 for schizophrenia and bipolar disorder in Han Chinese. Transl Psychiatry 6: e701. 10.1038/tp.2015.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Michelson Y, Freger V, Avraham Z, Venken KJT, Bellen HJ, Justice MJ, Wides R. 2011. Drosophila Ten-m and filamin affect motor neuron growth cone guidance. PLoS One 6: e22956. 10.1371/journal.pone.0022956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.