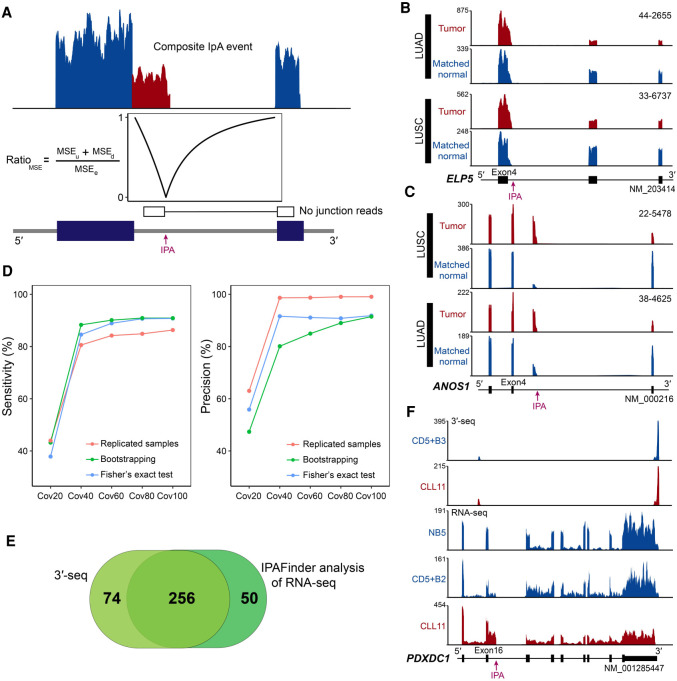
Figure 1.
Overview of the IPAFinder algorithm and evaluation of its performance. (A) Schematic diagram of IPAFinder in detecting composite terminal exon IpA event. Intronic poly(A) site is determined based on the expected drop in read coverage downstream from the predicted poly(A) site. Alternative splicing events are excluded by recognizing junction-spanning reads. (B,C) Two examples of IPAFinder-identified dynamically changed IpA events from TCGA RNA-seq data. IpA usage of the ELP5 gene (B, composite IPA site) and ANOS1 gene (C, skipped IPA site) is increased in both lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) compared with that in matched normal tissues. Sample IDs are shown at the top right corner of the corresponding RNA-seq density plot. IPA site is indicated by a red arrow. (D) Performance of IPAFinder in detecting differentially used IPA sites in terms of sensitivity and precision. The number of TPs, FPs, and predefined true differentially used IPA sites (Ps) are used to calculate sensitivity (TP/P) and precision (TP/[TP + FP]). For each coverage level, we repeated 10 times to calculate the mean value of sensitivity and precision. For samples without replicates, two methods, including a bootstrapping-based method and Fisher's exact test–based method, were assessed. (E) Venn diagram comparison of the number of recurrent up-regulated IpA events identified by IPAFinder and those by 3′-seq using the same data from CLL and immune cell samples. (F) An example of dynamically changed IpA events (PDXDC1) between CLL samples and normal B cells detected by IPAFinder, which was absent in 3′-seq.