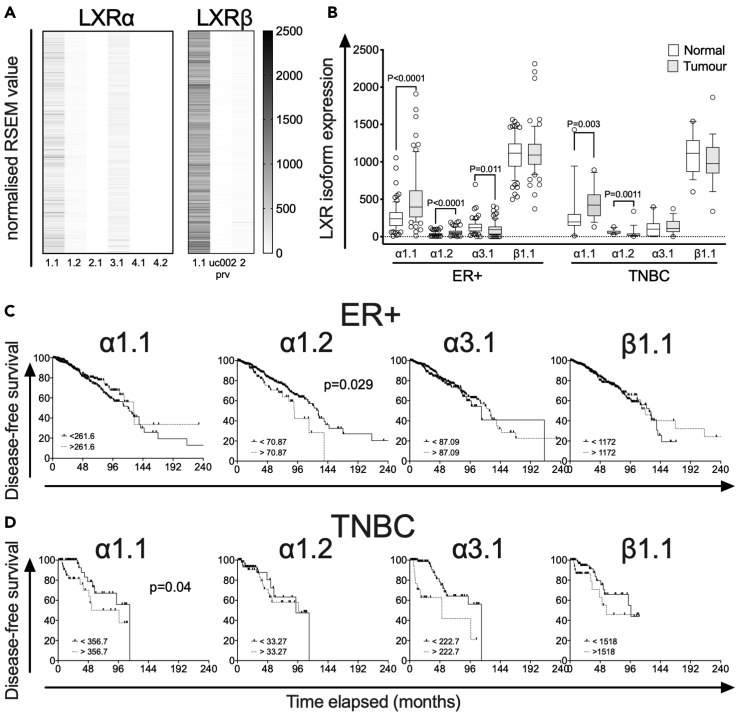
Figure 2.
LXR splicing events in BCa validated by TCGA Splicing Variants database (TSVdb)
(A) Heatmap visualization of normalized RSEM (RNA-Seq by Expectation Maximization) value expression of LXRα and LXRβ in 1,103 TCGA BCa samples. Note: Transcript uc002prv was only reported in TSVdb and is not recorded in NCBI, ENSEMBL, or UNIPROT databases.
(B) Comparison of LXR transcript variant expression, for α1.1, α1.2, α3.1, and β1.1, in matched tumor and normal tissues grouped by BCa subtype, ER+ (n = 78), and TNBC (n = 18). Statistical analysis by Mann-Whitney two-tailed U tests. P ≤ 0.05 was considered significant. The line shows the median value, the box shows 10th to 90th percentile, and whiskers show minimum-to-maximum value.
(C and D) Kaplan-Meier survival curves plotting disease-free survival of patients with TCGA BCa. The TCGA BCa samples (downloaded from TSvdb TCGA splicing database) were grouped by BCa subtype, (C) ER+ (n = 803), and (D) TNBC (n = 101). Survival curves of each subtype were separated into two groups: no event (ER+ = 675, TNBC = 79) and event (ER+ = 128; TNBC = 22) based on their overall survival data reported in cBioportal. p value is based on the log rank test, and data groups were considered significantly different if p < 0.05.