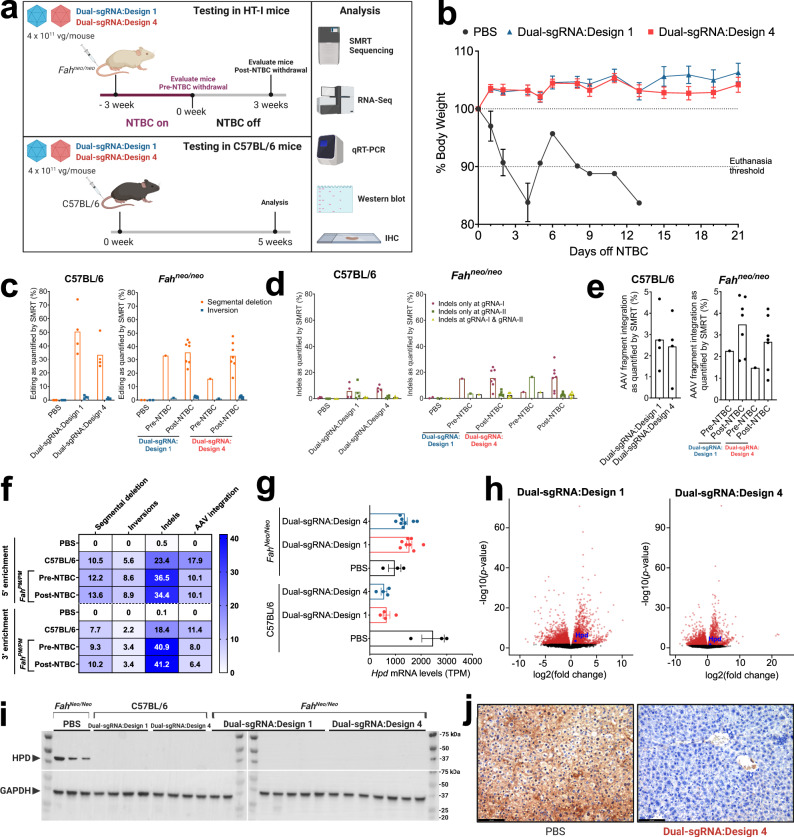
Fig. 2. In vivo gene editing using dual-sgRNA rAAV:Nme2Cas9 vectors.
a In vivo experimental plan to validate rAAV:Nme2Cas9 Dual-sgRNA:Designs 1 and 4 to disrupt the Hpd gene by tail vein injections of AAV8 vectors in adult Fahneo/neo and C57BL/6 mice. b Complete rescue of body weights after Dual-sgRNA:Designs 1 and 4 in Fahneo/neo mice. c Quantification of editing events in C57BL/6 (left) and Fahneo/neo (right) mice by SMRT sequencing analysis showing efficiencies of segmental deletion (orange) and inversion (blue) outcomes. d Bar graph showing the percentages of indels recorded in full-length (3.6 kb), UMI-corrected SMRT reads after Hpd editing by AAV8 delivery of Dual-sgRNA:Designs 1 and 4 in C57BL/6 (left) and Fahneo/neo (right) mice. The plot indicates indels recorded only at gRNA-I (in red), indels only at gRNA-II (in dark green), and indels at both gRNA-I and gRNA-II (yellow) as measured by SMRT sequence analysis. e rAAV fragment integration as detected by SMRT sequencing analysis. f Quantitation of editing events in UMI-corrected UDiTaS analysis reads after Hpd editing by AAV8 delivery of Dual-sgRNA:Design 4 in C57BL/6 and Fahneo/neo mice showing mean efficiencies of segmental deletion, inversion, and AAV fragment integration. g mRNA levels from RNA-seq (transcripts per million) of Hpd wild-type mRNA in the livers of C57BL/6 mice and Fahneo/neo mice. h Volcano plots showing differentially expressed genes with false discovery rate ≤5% after adjusting for multiple comparisons (red; analysis detailed in Methods) between Dual-sgRNA:Design 1 and PBS-treated Fahneo/neo mice (left) and Dual-sgRNA:Design 4 and PBS-treated Fahneo/neo mice (right). i Total HPD protein knockout as shown by anti-HPD Western blot using total protein collected from mouse liver homogenates. This analysis of samples from multiple mice was performed once. j Representative images of immunostaining for HPD in liver tissues in Fahneo/neo mice injected with PBS (left) or Dual-sgRNA:Design 4 (right). Scale bar is 100 μm. All mice in the cohort were examined once by this method (see Supplementary Fig. 2). Data were presented as mean values ± s.e.m. Sample size in panels b–g: (n = 3 PBS-injected C57BL/6 mice; n = 4 Dual-sgRNA.Design 1 or 4 injected C57BL/6 mice; n = 1 in pre-NTBC Fahneo/neo cohort; n = 7 in post-NTBC Dual-sgRNA.Design 1 or 4 injected Fahneo/neo cohort).