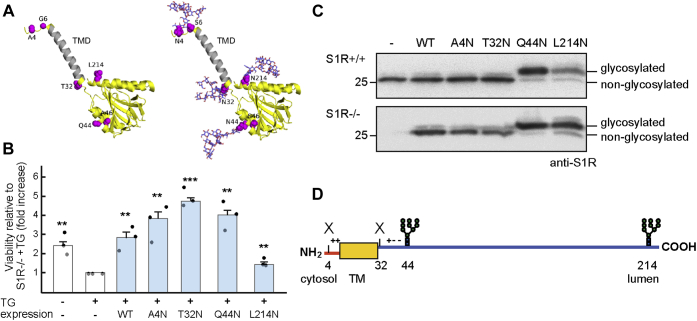
Figure 4.
An approach introducing N-glycosylation sites into untagged S1R indicates a type II membrane topology.A, 3D structure of the human S1R (left) and of the same structure showing potential N-glycans (Man9GlcNAc2) (shown as sticks) (right), at glycosylation sites introduced into four different constructs. The protein is shown as a yellow ribbon, with the positions targeted for mutations (4, 6, 32, 44, 46, and 214) shown as magenta spheres and labeled. The transmembrane domain α-helix (TMD) is colored in gray. The N-terminus of the protein is on the top. B, S1R−/− HEK293 cells, transfected with WT S1R or with the N-glycosylation site constructs, mutations in parenthesis: A4N (A4N and G6S), T32N (T32N), Q44N (Q44N and A46S), and L214N (L214N) or mock transfected were treated with 2 μg/ml TG for 24 h and cell viability was tested compared with the treated untransfected cells using a RealTime-Glo MT cell viability assay. The graph represents the mean of three independent experiments ±SEM, p values (compared with treated untransfected cells) ∗∗ <0.01, ∗∗∗ =0.00015. Student's t test (paired, two-tailed). C, S1R+/+ (top) or S1R−/− (bottom) HEK293 cells were transfected with WT S1R or with the N-glycosylation site constructs or left untransfected and subjected to immunoblotting with an anti-S1R antibody. D, scheme of the results of the experiment in (C). The N(44)LS and N(214)TT sites were glycosylated, while the other two introduced sites, N(4)VS and N(32)QS, were not, suggesting a type II membrane protein topology. Charges in amino acids in the juxtamembrane region are shown.