Figure 7 .
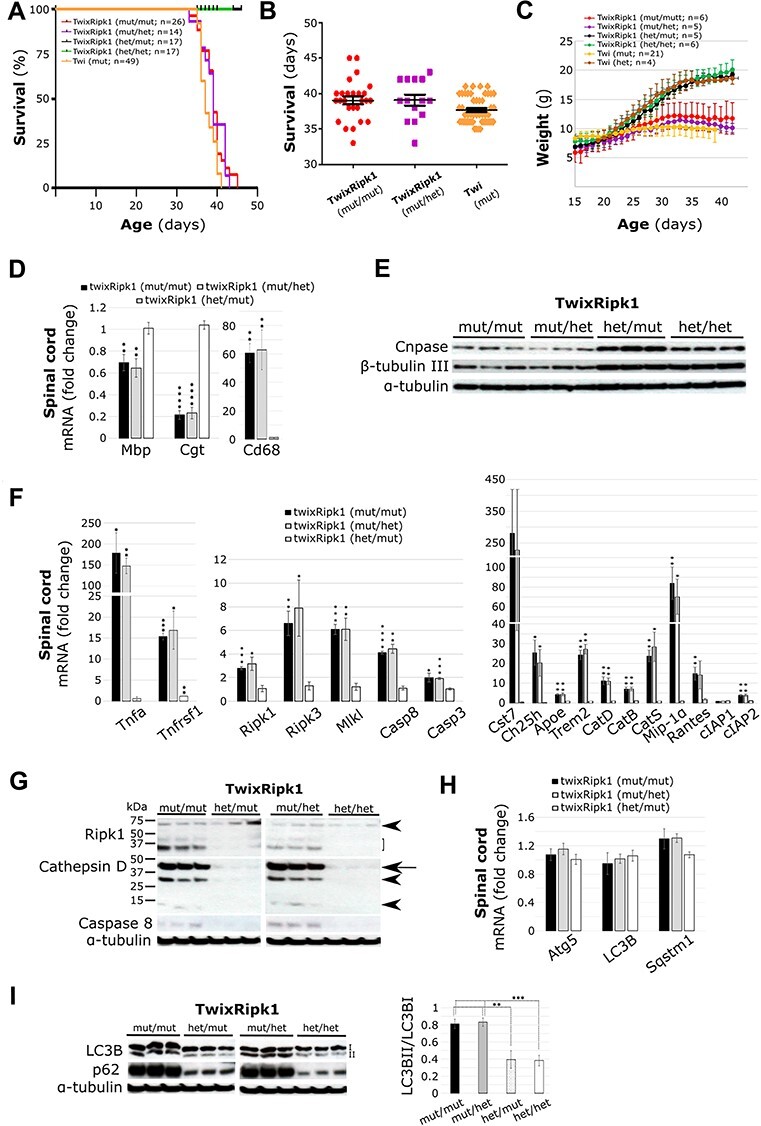
Genetic inhibition of Ripk1 kinase activity neither extends life nor prevents disease progression or pathological features in twitcher. (A, B) Survival of mutant twi-2J (twi) which is wild type for Ripk1 does not differ from twi homozygous or heterozygous for the K45A Ripk1 mutation, and thus, no survival benefit was accrued. Kaplan–Meier (A) and scatter plot with mean ± SEM (B). (C) Mice were weighed daily from age 15 days until they reached the HEP and mean ± SD was plotted. The K45A Ripk1 mutation whether in homo- or heterozygosity does not alter the weight of heterozygous or mutant twitchers. (D) Myelin defects and gliosis was assessed by RT-qPCR of total RNA in the spinal cord HEP with markers: Mbp (Myelin basic protein), Cgt (UDP-galactose:ceramide galactosyl-transferase), and Cd68 (Cluster of differentiation 68); (E) Myelin and axon protein content was examined in sciatic nerve with markers Cnpase (2′,3’-Cyclic Nucleotide 3’-Phosphodiesterase) and β-tubulin III, respectively by immunoblotting at HEP. (F) Evaluation of expression of core ripoptosome/necroptosome components, neuroinflammation and DAM genes was performed by RT-qPCR of total RNA from HEP spinal cord with: Tumour necrosis factor (Tnf), Tumour necrosis factor receptor (Tnfrsf1), Receptor-interacting serine–threonine kinase 1 (Ripk1), Receptor-interacting serine–threonine kinase 3 (Ripk3), Lineage kinase domain-like (Mlkl), Caspase-8 (Casp8), Caspase-3 (Casp3); Cystatin F (Cst7), Cholesterol 25-hydroxylase (Ch25h), Apolipoprotein E (Apoe), Triggering receptor expressed on myeloid cells 2 (Trem2), Cathepsin D (CatD), B (CatB), S (CatS), Macrophage inflammatory protein 1 alpha (Mip-1α), Regulated on activation normal T cell expressed and secreted (Rantes), Cellular Inhibitor of Apoptosis Protein 1 (cIAP1) and 2 (cIAP2). (G) Immunoblotting of Ripk1, cathepsin D and caspase 8 from brain stem at HEP. Ripk1 species full-length (arrowhead), cleaved forms (bracket); catepsin D intermediate (arrow) and mature forms (arrowhead); caspase-8 (uncleaved). Autopohagy was examined by (H) RT-qPCR of total RNA from HEP spinal cord with markers Atg5 (autophagy related 5), LC3B (microtubule-associated protein 1 light chain 3 beta), and Sqstm1 (sequestosome 1 codes for p62); (I) Immunoblotting of brain stem with LC3BII and p62, and ratio LC3BII/LC3B1 relative to α-tubulin. RT-qPCR RNA quantification is given relative to healthy controls (TwixRipk1: het/het), matched for age and sex. Ten microgram protein homogenate was loaded per lane in B, and 100 μg for D and F. Blots were stripped of first antibody, and re-re-probed with subsequent antibodies. Mutant (mut), heterozygote (het). Student’s t-test; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
