Figure 3. Inactivation of Rev3l impairs expression of numerous developmentally and imprinted genes.
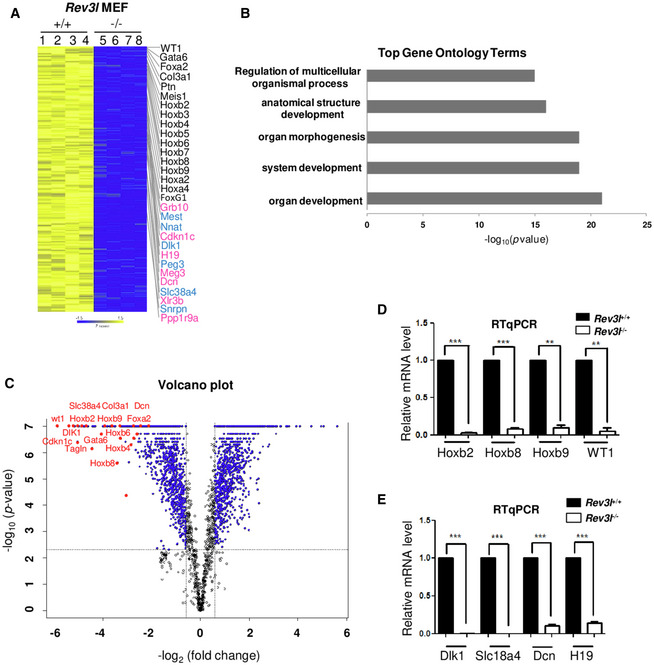
- Heat map showing log2 fold change in differentially expressed genes in Rev3l +/+ and Rev3l −/− MEFs from 2 independent biological experiments (Rev3l+/+: samples 1 and 2, Rev3l−/−: samples 5 and 6) with 2 technical replicates (samples 3,4,7,8: technical repeats from samples 1,2,5,6, respectively). Several developmentally regulated genes and imprinting genes (paternally in blue and maternally in pink) down‐regulated in Rev3l −/− MEFs are indicated on the right. Yellow and blue indicate high and low mRNA expression levels, respectively.
- Top five biological process gene ontology (GO) terms of genes found down‐regulated in Rev3l −/− MEFs transcriptome analysis.
- Volcano plot shows in red genes involved in development and imprinting with high fold changes ≥ 3 (FDR < 0.05).
- Relative mRNA levels of four genes involved in development (Hoxb2, Hoxb8, Hoxb9, and WT1) were validated through qRT–PCR from Rev3l +/+ and Rev3l −/− MEF samples. The data were normalized to the amount of GAPDH mRNA. Error bars indicate standard error of the mean for three independent experiments. ***P < 0.001 and **P < 0.005 by Student’s t‐test.
- Relative mRNA levels of genes involved in imprinting (paternal expressed genes: Dlk1 and Slc18a4 and maternal expressed genes: Dcn1 and H19) were validated through qRT–PCR from Rev3l +/+ and Rev3l −/− MEF samples. The data were normalized to the amount of GAPDH mRNA. Error bars indicate standard error of the mean for three independent experiments. ***P < 0.001 by Student’s t‐test.
Source data are available online for this figure.
