Figure 2. STm effector-host target physical interactions in RAW264.7 macrophages.
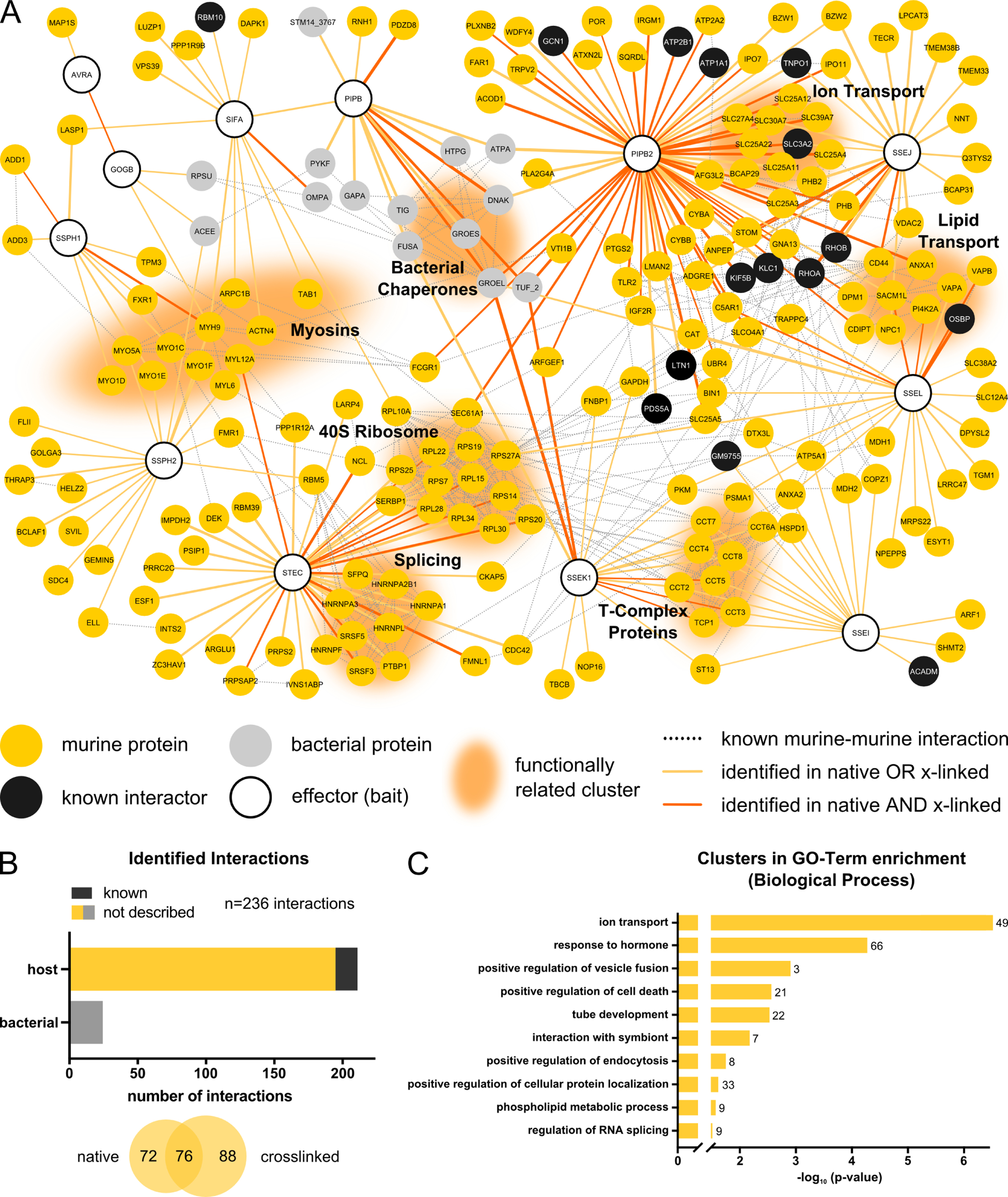
(A) Network of PPIs identified between 12 STm effectors and their target proteins in RAW264.7 cells at 20 hpi. Only effectors identified as bait in AP-QMS and target proteins passing the criteria described in Fig. 1B are shown here. Host proteins from RAW264.7 cells are shown in gold or black (previously identified interactions; Table S3). STm are in grey. The edge color denotes the conditions interaction captured, and the thickness is proportional to the fold change (Log2). Functionally related clusters are grouped and annotated accordingly. Network was generated using Cytoscape version 3.7.2 (Shannon et al., 2003). Murine-murine, as well as bacterial-bacterial functional interactions were extracted from the built-in STRING DB version 11 (Szklarczyk et al., 2019) protein query for Mus musculus and Salmonella with a confidence cutoff of 0.7.
(B) Overview of identified PPIs in RAW264.7 cells at 20 hpi. Hits are grouped according to whether they are of murine or STm origin (upper histogram), or according to whether they were detected in native or cross-linked pulldown samples (lower Venn diagram).
(C) GO-term analysis for enriched processes among all identified PPI partners. GO-term clusters are ordered according to the enrichment significance (negative logarithmic, corrected for multiple testing) (Benjamini and Hochberg, 1995; Bindea et al., 2009) and top 10 GO-clusters are displayed. n signifies the number of proteins present in cluster. Enrichments were normalized to the combined background proteome from AP-QMS experiments. Full enrichments lists can be found in Table S4.
