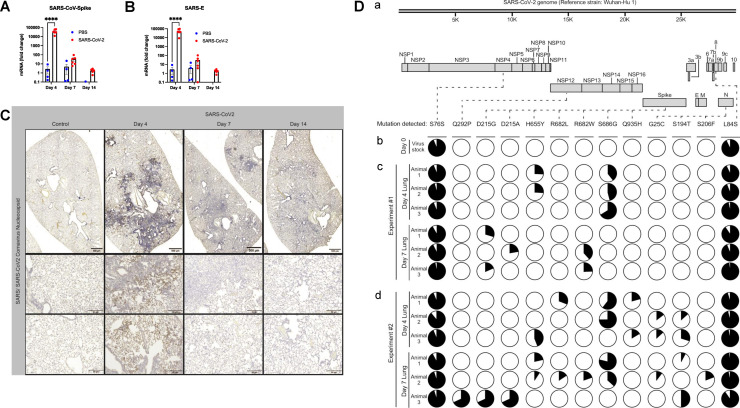
Fig 1.
SARS-CoV-2 inoculation leads to infection in Syrian golden hamsters. (A,B) Total RNA was harvested from hamster lung tissue, followed by analysis by qPCR. SARS-CoV-2 infected hamsters demonstrated significantly increased SARS-CoV-2-spike and SARS-E mRNA at 4 dpi but no significant differences at 7 or 14 dpi. n >= 4 hamsters and/or group. Bar graph represents mean fold change ± SEM. Dots represent individual animals. ****P < 0.0001 as determined by ANOVA with post-hoc Sidak's multiple comparisons test. (C) Representative images from hamster lung tissue demonstrate increased staining for SARS-CoV-2 spike protein by immunohistochemistry (IHC) at 4 dpi but no significant increased viral protein at 7 or 14 dpi. (D) SARS-CoV-2 intra-host sequence analysis. (Da) Depiction and annotation of the SARS-CoV-2 genome, as modified from Gordon et al.40 to show the locations of thirteen mutations detected in (Db) viral RNA extracted from the SARS-CoV-2 stock used to infect all the hamsters in this study, (Dc) viral RNA extracted from individual hamster lungs at post-infection Day 4 or post-infection Day 7 (n = 3 hamsters and/or time point), as collected during one experiment (Experiment #1), and (Dd) viral RNA extracted from individual hamster lungs at the same time points, as collected during a second independent experiment (Experiment #2). From left to right, these mutations were in the following genes and nucleotide positions (protein changes are indicated in parentheses): NSP4.228C→T (S76S), NSP12.875A→C (Q292P), Spike.644A→G (D215G), Spike.644A→C (D215A), Spike.1963C→T (H655Y), Spike.2044C→T(R682W), Spike.2045G→T(R682L), Spike.2056A→G(S686G), Spike.2805A→T(Q935H), N.73G→T(G25C), N.580T→A(S194T), N.617C→T(S206F), and ORF8.251T→C(L84S). The pie charts under each mutation indicate the proportion of reads in a given sample that either contained the mutation (the black portion of the pie) or wild type nucleotide (the white portion of the pie).