FIGURE 5.
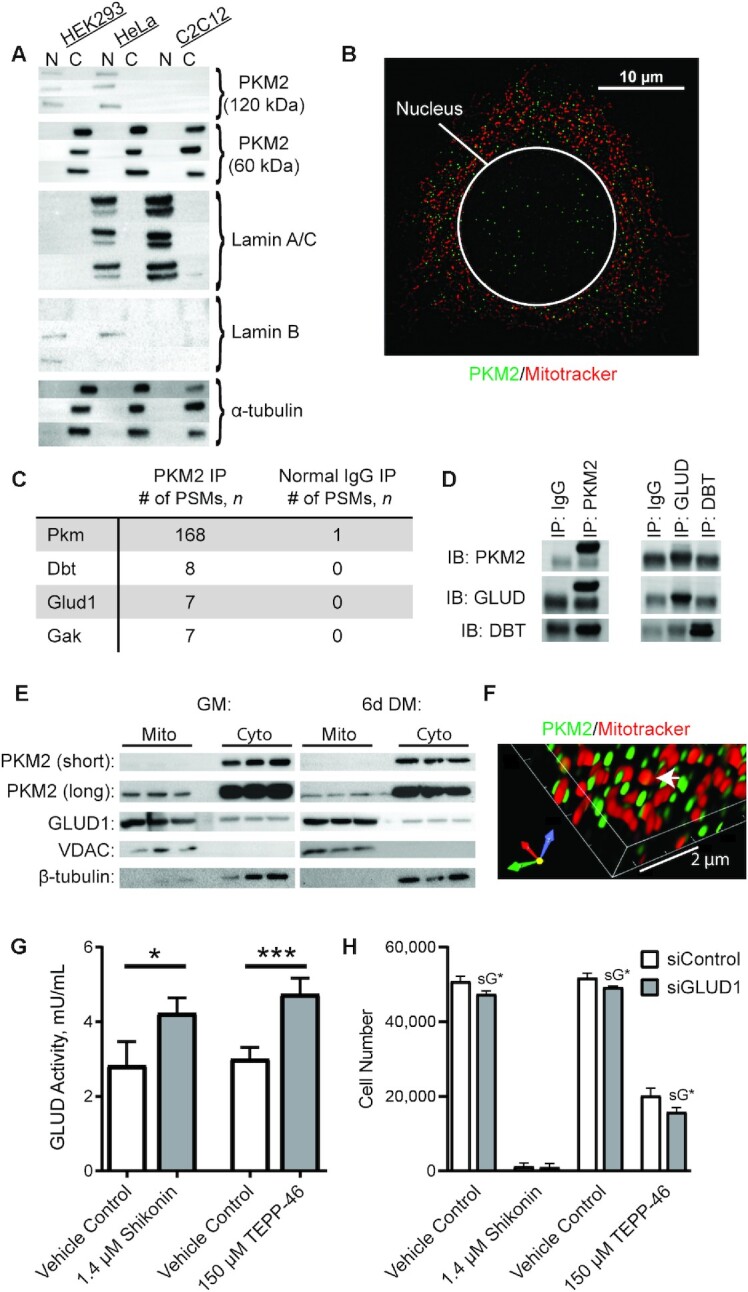
PKM2 localization and binding partners in MPCs. (A) Immunoblotting of PKM2 and cytosolic (tubulin) and nuclear (Lamin A/C and lamin B) markers in MPCs, HeLa cells, and HEK293 cells. N indicates nuclear fraction and C indicates rest of cell (n = 3). (B) Confocal microscopy of Hoechst, mitotracker, and PKM2 in MPCs. (C) Top 3 candidate PKM2 interacting proteins identified using nontargeted MS-based proteomic analysis. (D) Validation of PKM2 binding partners by IP coupled to immunoblotting. (E) Subcellular fractionation of cytosolic and mitochondrial fractions (n = 3 replicates). (F) Co-staining of PKM2 and Mitotracker imaged using structured illumination microscopy. (G) GLUD activity in vehicle control, shikonin, or TEPP-46 treated cultures (n = 4). (H) Cell number in cultures treated with vehicle control, shikonin, or TEPP-46 and siControl or siGLUD1 (n = 4). Values are mean ± SD. *P < 0.05, ***P < 0.001 (asterisks indicate significant effects). sG*, effect of siGLUD1. Dbt, dihydrolipoamide branched-chain transacylase E2; DM, differentiation medium; Gak, cyclin G associated kinase; Glud1, glutamate dehydrogenase 1; GM, growth medium; IB, immunoblot; IP, immunoprecipitation; MPC, muscle progenitor cell; Pkm, pyruvate kinase; PKM2, pyruvate kinase M2; PSM, peptide spectral match; siControl, small interfering RNA non-targetting control; siGLUD1, small interfering RNA targetting GLUD1; VDAC, voltage-dependent anion channel.
