Figure 1.
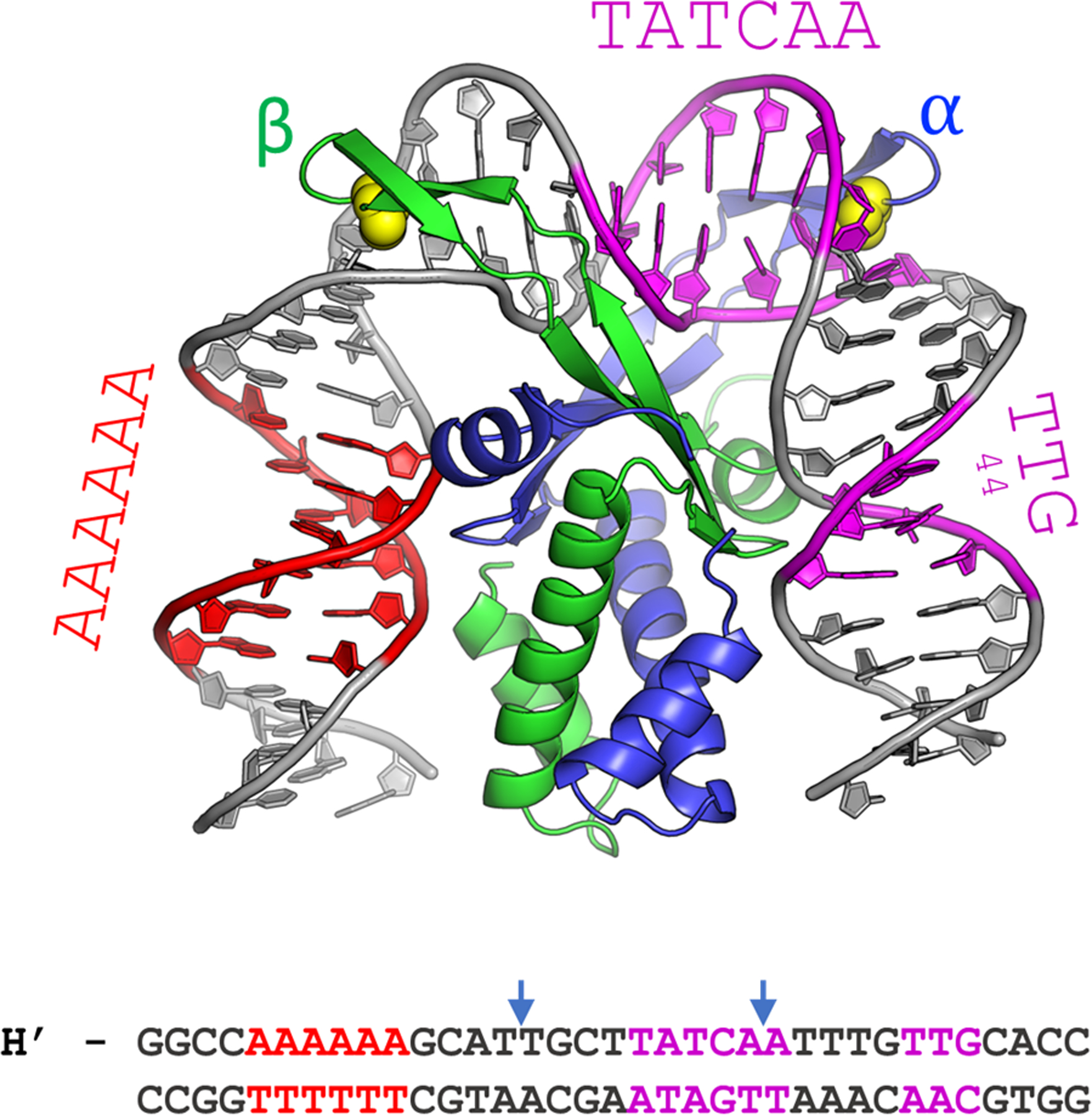
The cocrystal structure of IHF in complex with the H’ binding site from phage lambda. The α- and β-chains of the IHF protein are shown in blue and green, respectively, with the conserved proline residues shown as yellow spheres. The DNA is shown in gray, with the consensus region highlighted in magenta and the A-tract in red. The sequence shown below is that of the 35-mer that contains the H’ binding site, which was used to obtain the cocrystal structure with IHF. In the complex, the DNA is sharply kinked at the two sites indicated by the blue arrows. In the DNA oligomer used for the structural studies, the DNA was nicked at a position shifted 1 bp to the 3’-side of the left blue arrow, to facilitate crystal packing. That nick was “sealed” in silico to generate the model shown (starting from PDB ID 1IHF).
