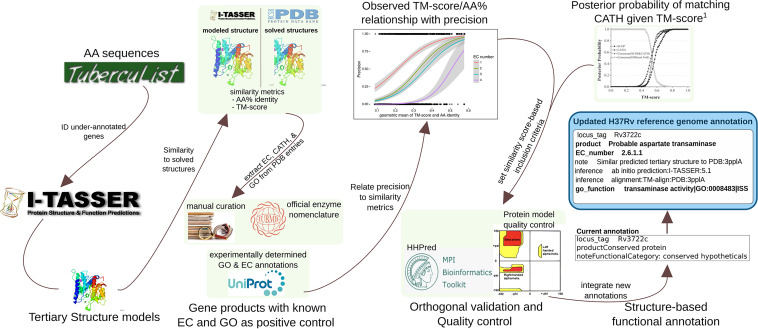
FIG 2.
Information flow for producing annotations from structural similarity. The flow of information and procedures for acquiring, processing, filtering, and representing information, running from retrieval of amino acid sequences to the final updated H37Rv annotation. Some details are omitted for clarity. The 1,725 amino acid sequences were retrieved from TubercuList and run through a local installation of I-TASSER v5.1. Of 1,725 amino acid sequences, 1,711 had models generated successfully. Comparison metrics for sequence (amino acid identity) and structure (TM-score) were extracted from I-TASSER output. To set criteria for annotation transfer, precision (equation 1) of GO Term and EC number concordance between similar matches on PDB and true function of 363 positive controls with GO terms and EC numbers of known function were regressed against extracted similarity metrics to generate a curve relating the geometric mean of TM-score and amino acid similarity to precision. These informed inclusion thresholds for transferring GO and EC annotations from structures on PDB similar to the 1,711 modeled structures. CATH topology folds were transferred according to a previous precision curve based on TM-score. This threshold was also used for inclusion of protein classes that vary in sequence more than structure (e.g., transporters) and as criteria for transferring annotations from structures that were not annotated with EC numbers or GO terms. Annotations derived only from structure were passed through orthogonal validation and manual structure analysis for verification that transferred annotations were reasonable. All annotations were programmatically collated into an updated H37Rv reference genome annotation.