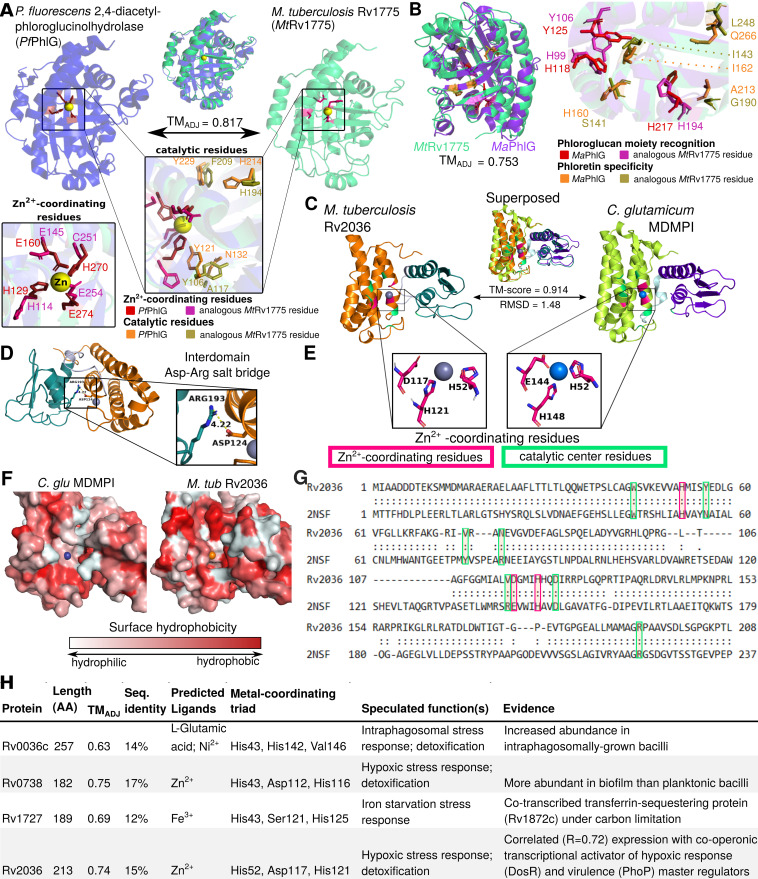
FIG 4.
Manual structural analysis refines functional annotations uncorroborated by HHpred. (A and B) Conservation of structure and sequence features essential for C—C bond hydrolysis supports the inferred hydrolase function of Rv1775. (A) Structural alignment of modeled Rv1775 and its closest structural match (PDB ID 3hwp), a 2,4-diacetyl-phloroglucinolhydrolase of Pseudomonas fluorescens (PfPhlG), The structures are superposed (top). Zoomed and reoriented images of PfPhlG zinc-coordinating (box on left) and catalytic (popout) residues superposed with analogous MtRv1775 residues. (B) Comparison of functional and structural features between MtRv1775 and a putative PhlG homolog of M. abscessus (MaPhlG), phloretin hydrolase, which catalyzes C—C bond hydrolysis of a different substrate. Comparison carried out in a similar scheme as in panel A. Superposition of the putative homologs, color annotated with conserved residues essential for phloroglucol moiety recognition and for phloretin substrate specificity in MaPhlG (47). The structural similarity and conserved zinc-coordinating and catalytic residues affirm Rv1775 as a bona fide C—C hydrolase, potentially with a substrate that includes a phloroglucol moiety but likely not phloretin. Conservation of structure and sequence features characteristic of DinB-like metalloenzymes exemplified by structural homology of Rv2036 and a mycothiol-dependent maleylpyruvate isomerase from Corynebacterium glutamicum (C. glu MDMPI) (C to G). (C) Superposition of Rv2036 structure model and C. glu MDMPI (PDB ID 2nsf). Conserved Zn2+-coordinating (pink) and catalytic (green) residues are highlighted. (D) Highly conserved residues Arg222 (C-terminal domain, Arg193 in Rv2036) and Asp151 (N-terminal domain, Asp124 in Rv2036) are in close proximity (4.22 Å), suggesting conservation of their proposed role as interdomain protein stabilizers (51). (E) Spatial conservation of Zn2+-coordinating residues of the catalytic triad (Asp and Glu are observed interchangeably) is consistent with conserved catalytic function. (F) Surface hydrophobicity of Rv2036 model and 2nsf shows that the hydrophilic core proposed to underlie MDMPI catalysis (51) is relatively conserved. (G) Structure-based sequence alignment of Rv2036 and C. glu MDMPI with conserved residues was manually annotated according to prior work (51). (H) Summary of relevant genomic context potentially informative of function, protein similarity metrics between putative M. tuberculosis MDMPI homologs and C. glu MDPMI, and predicted protein features. All structural images were rendered in PyMOL. Structurally homologous sequence alignments are based on TM-align (22) (**, <5 Å between residues; *, <10 Å between residues).