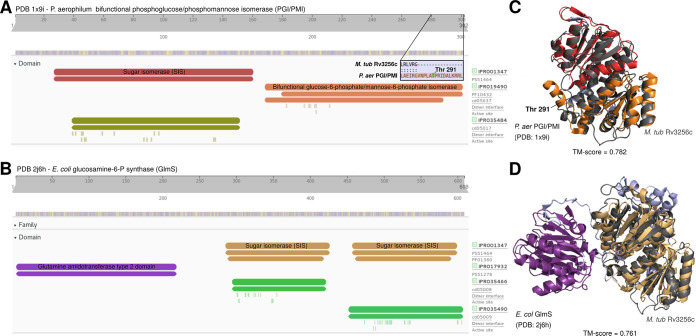
FIG 8.
Structural analysis informs specific functional hypotheses for the basis of PZA resistance in the Rv3256c::Tn mutant. (A to D) Structural analysis identifies Rv3256c as a sugar isomerase (SIS) domain-containing protein likely involved in phosphosugar metabolism or its regulation. InterPro functional domains are displayed for the two strongest structural matches of Rv3256c, Pyrobaculum aerophilum bifunctional phosphoglucose/phosphomannose isomerase (P. aer PGI/PMI) (A) and Escherichia coli glucosamine-6-P synthase (E. col GlmS) (B). The InterPro domains labeled in panels A and B are mapped onto the three-dimensional (3D) structures of P. aer PGI/PMI (C) and E. col GlmS (D). Rv3256c (charcoal) modeled protein structure is optimally superposed on each of its matches. Rv3256c is structurally homologous to the SIS domains of E. col GlmS and P. aer PGI/PMI and exhibits the alpha-beta-alpha sandwich fold of SIS (128). The popout in panel A (**, <5 Å between residues) and labeled residue in panel C show the threonine residue essential for isomerase activity in P. aer PGI/PMI (Thr291) and other PMI homologs. The Thr291-containing region appears to be absent from Rv3256c. Likewise, Rv3256 lacks a glutamine amidotransferase domain homologous to E. col GlmS. From this structural evidence, we conclude that Rv3256c is a SIS domain protein putatively involved in phosphosugar metabolism and/or its regulation. Structural images were rendered in PyMOL. Structurally homologous sequence alignments from TM-align (22).