FIG 9.
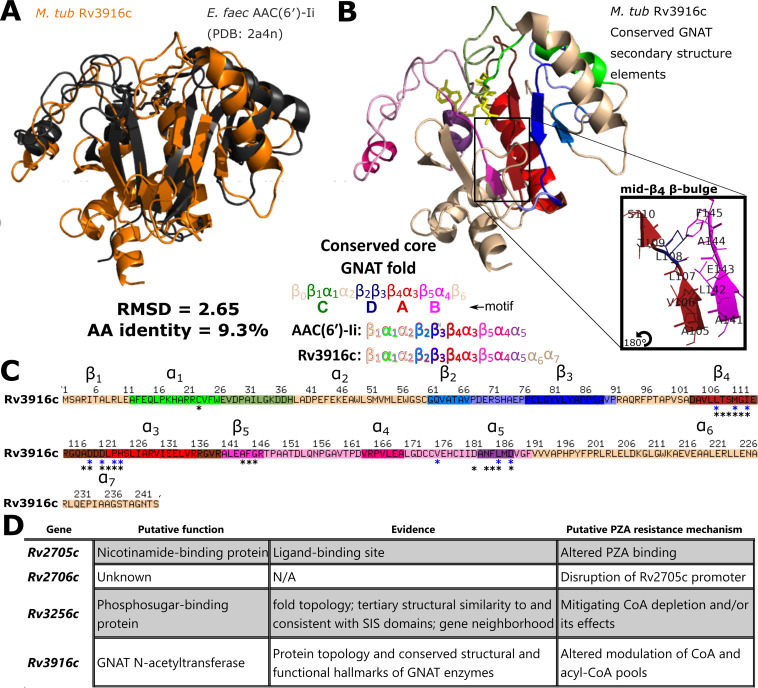
Structural analysis of transposon mutants refines functional hypotheses for their role in PZA resistance. (A to C) Structural analysis supports Rv3916c as a general control nonrepressible 5 (GCN5)-related N-acetyltransferases (GNAT). Rv3916c matches exclusively comprised GNAT enzymes with low sequence similarity, typical among homologous GNAT enzymes (129). (A) Rv3916c (orange) superposed with its closest structural match (PDB ID 2a4n, charcoal), Enterococcus faecium aminoglycoside 6′-N-acetyltransferase [E. faec AAC(6′)-Ii]. RMSD, root mean square deviation. (B) M. tuberculosis Rv3916c structural model (top) and secondary structure topology (bottom), colored according to the four core conserved GNAT folds (wheat = poorly conserved secondary structure element) and secondarily (tones of the primary colors) by secondary structure elements in E. faec AAC(6′)-Ii (as defined by reference 130) with which Rv3916c structurally aligns. All secondary structure elements of E. faec AAC(6′)-Ii are present in Rv3916c, with two gratuitous alpha-helices in its C-terminal arm (which is not well conserved among GNAT enzymes [129]). The distinctive β-bulge (popout) within the β4 strand (red; bulge residues colored blue) characteristic of GNAT enzymes is present in Rv3916c, diverting β4 away from β5 to create the chasm where the acetyl-coenzyme A (yellow) is predicted to bind. (C) Primary sequence of Rv3916c colored according to the scheme described in panel B. Asterisks mark predicted acetyl-CoA binding residues (black) and residues structurally aligning to known CoA-interacting residues (blue). All known CoA-interacting residues from E. faec AAC(6′)-Ii are conserved in Rv3916c, and all predicted acetyl-CoA binding sites coincide with or directly flank demonstrated sites of CoA interaction. The presence of the features in Rv3916c suggests it is a GNAT enzyme. GNAT enzymes catalyze transfer of an acyl moiety from an acyl-CoA to various substrates (129), making Rv3916c a probable acyl-CoA acetyltransferase and implicating PRv3916c::Tn in the CoA pool modulation model of PZA resistance. (D) Summary of functional hypotheses for PZA resistance-conferring transposon mutants. All structural images were rendered in PyMOL. Structurally homologous sequence alignments are based on TM-align (22). N/A, not available.