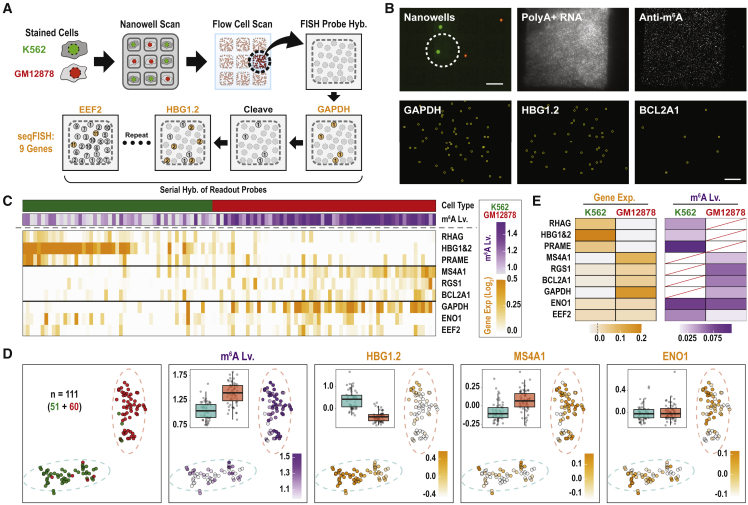
Figure 3.
Application of seqFISH on the single-cell m6A assay
(A) Experimental workflow. A mixture of K562 (SYTO9, green) and GM12878 (SYTO87, red) cells was applied to the nanowell plate. The nanowell plate and flow cell were scanned, as described in Figure 2A. Gene-specific primary probes were hybridized to nine targeted mRNAs. To quantify each targeted transcript, we sequentially hybridized secondary readout probes, removing the fluorophores between rounds by disulfide cleavage.
(B) Top left: representative nanowell scan. Scale bar, 100 μm. Remaining images show polyA+ RNA; m6A-modified RNA; and seqFISH for GAPDH, HBG1 and 2, and BCL2A1 from a single well. Scale bar, 20 μm.
(C) Heatmap showing m6A levels and relative expression levels of nine target genes at a single-cell level in K562 (green) and GM12878 (red) cells.
(D) tSNE visualization of 48 qualified single-cell seqFISH profiles. Dashed circles indicate clusters (subpopulations). Single cells are colored according to cell type on the nanowell array (green or red), m6A level (purple), or relative gene expression levels (orange). Box plots show the m6A level (m6A/total transcripts) or log-mean gene expression levels among clusters.
(E) Heatmap of gene expression levels and gene-specific m6A levels in K562 and GM12878 cells. Red diagonal lines represent “not applicable” values with log-mean gene expression levels less than −0.02.
See also Figure S3 and Table S2.