The molecular basis for CRAC channel regulation by the SOCE-associated regulatory factor (SARAF) remains insufficiently understood. Zomot, Achildiev et al. present a new role for SARAF in the activation of STIM1 and subsequent coupling to the Orai1 channel.
Abstract
Store-operated calcium entry (SOCE) through the Ca2+ release–activated Ca2+ (CRAC) channel is a central mechanism by which cells generate Ca2+ signals and mediate Ca2+-dependent gene expression. The molecular basis for CRAC channel regulation by the SOCE-associated regulatory factor (SARAF) remained insufficiently understood. Here we found that following ER Ca2+ depletion, SARAF facilitates a conformational change in the ER Ca2+ sensor STIM1 that relieves an activation constraint enforced by the STIM1 inactivation domain (ID; aa 475–483) and promotes initial activation of STIM1, its translocation to ER–plasma membrane junctions, and coupling to Orai1 channels. Following intracellular Ca2+ rise, cooperation between SARAF and the STIM1 ID controls CRAC channel slow Ca2+-dependent inactivation. We further show that in T lymphocytes, SARAF is required for proper T cell receptor evoked transcription. Taking all these data together, we uncover a dual regulatory role for SARAF during both activation and inactivation of CRAC channels and show that SARAF fine-tunes intracellular Ca2+ responses and downstream gene expression in cells.
Introduction
Store-operated calcium entry (SOCE) represents a key mechanism by which cells generate Ca2+ signals and maintain Ca2+ homeostasis by replacing Ca2+ lost from the ER with Ca2+ that enters the cytoplasm through plasma membrane (PM) channels (Putney, 1986). In some cell types, members of the transient receptor potential channel (TRPC) family were shown to contribute to SOCE, but, in most cells, SOCE is mediated by Ca2+ release–activated Ca2+ (CRAC) channels (Ambudkar et al., 2017; Lunz et al., 2019). The physiological role of CRAC channels is particularly well understood in T lymphocytes, where calcium signaling is essential for various aspects of T cell–mediated immunity (Trebak and Kinet, 2019).
The primary components of CRAC channels are stromal interaction molecule 1 (STIM1), the Ca2+ sensor of the ER, and the PM channel protein calcium release-activated calcium channel protein 1 (Orai1). STIM1 activates Orai1 channels through a short STIM-Orai–activating region (referred to either as SOAR [aa 344–442] or CAD [aa 342–448]), which is necessary and sufficient for the interaction with Orai1 (Yuan et al., 2009; Park et al., 2009). In the resting state of STIM1, the SOAR is masked by intramolecular interactions with other regions in STIM1 and does not freely interact with Orai1 (the term “SOAR” is used hereafter for simplicity). Shielding of SOAR from Orai1 is achieved by a well-characterized intramolecular interaction between SOAR and the STIM1 coiled-coil 1 region (CC1; aa 238–344; Korzeniowski et al., 2010; Muik et al., 2011; Yang et al., 2012) and by a second inhibitory mechanism that involves a region located C-terminally to SOAR (Jha et al., 2013). The STIM1 inactivation domain (ID; aa 470–491) was recently identified as a key element responsible for the latter inhibitory interaction that maintains SOAR in a resting conformation (Lee et al., 2019). The transition of STIM1 from resting to active state involves several steps, some of which are becoming well understood. Depletion of Ca2+ from the ER causes Ca2+ release from the lumen-facing STIM1 EF-hand Ca2+ binding domain and elicits dimerization of adjacent SAM domains (Liou et al., 2005; Stathopulos et al., 2006). These changes drive an extensive conformational change in the cytosol-facing C-terminal region (Muik et al., 2011; Stathopulos and Ikura, 2010; Fahrner et al., 2014; Zhou et al., 2013). This conformational change is aided by the STIM-activating enhancer STIMATE, which binds to the CC1 and facilitates homomerization of CC1 domains and exposure of SOAR (Jing et al., 2015). The mechanism by which SOAR is freed from inhibition imposed by the STIM1 ID remains elusive. The extension of the cytosol-facing segment of STIM1 elicits recruitment to specialized ER regions found close to the PM, where STIM1 interacts first with PM phosphatidylinositol bisphosphate or trisphosphate lipids to form ER–PM junctions and subsequently with Orai1 channels to initiate Ca2+ entry.
The rise in cytoplasmic Ca2+ stimulates various calcium-dependent signaling cascades and feedback to suppress channel activity by fast Ca2+-dependent inactivation (FCDI; τ = ∼10–20 ms) and slow Ca2+-dependent inactivation (SCDI; τ = ∼1–3 min) mechanisms. In a previous study, we identified SOCE-associated regulatory factor (SARAF), a single-pass ER membrane protein, as a regulator of cellular calcium homeostasis (Palty et al., 2012). Following ER Ca2+ depletion, SARAF migrates to ER–PM junctions, where STIM1-Orai1 puncta are formed. Structure–function studies have shown that SARAF contributes to SCDI through its cytosol-facing domain and that this activity is modulated by the oligomeric state of its luminal domain and by recruitment of STIM1-Orai1 channel complex to ER–PM junctions enriched with phosphatidylinositol 4,5-bisphosphate (Palty et al., 2012; Kimberlin et al., 2019; Maléth et al., 2014). SARAF interacts with STIM1 through binding to SOAR, and deletion mutations within the C-terminal inhibitory domain (CTID; residues 448–530) of STIM1 were shown to regulate this interaction (Jha et al., 2013). Curiously, however, the interaction between STIM1 and SARAF increases following ER Ca2+ depletion and under low levels of cytosolic Ca2+ that do not elicit SCDI (Jha et al., 2013; Palty et al., 2012; Maléth et al., 2014). Importantly, although changes in STIM1–SARAF interaction precede the induction of SCDI and correlate with development of the CRAC current (ICRAC), it is not known whether SARAF affects CRAC channel activation.
In the present study, we abolished SARAF expression in several types of cells and employed electrophysiological, biochemical, and fluorescence microscopy techniques to investigate the role of SARAF in the regulation of CRAC channel activation and downstream signaling. We found that upstream from its effect on SCDI, SARAF initially promotes ICRAC development by releasing an activation constraint imposed by the STIM1 ID. We further reveal that in T lymphocytes, the contribution of SARAF to CRAC channel activation plays a key role in T cell receptor (TCR)-mediated Ca2+-dependent nuclear factor of activated T cells (NFAT) transcription. Collectively, these results demonstrate that through dual modulation of CRAC channel function, SARAF fine-tunes intracellular Ca2+ levels and downstream signaling in cells.
Results
SARAF affects both size and inactivation of ICRACs
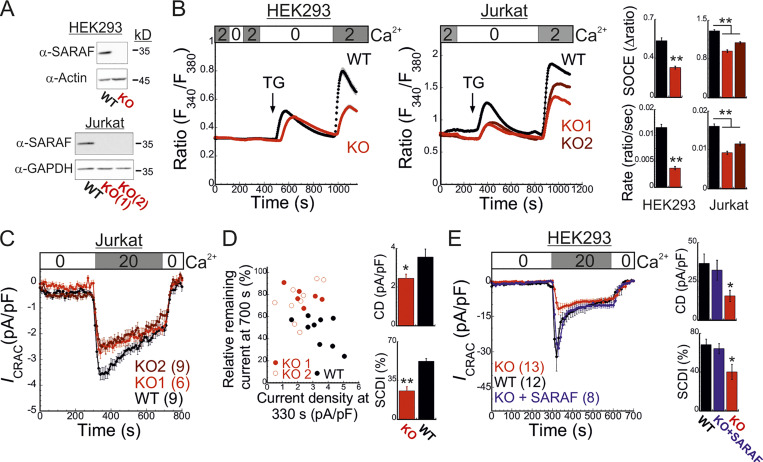
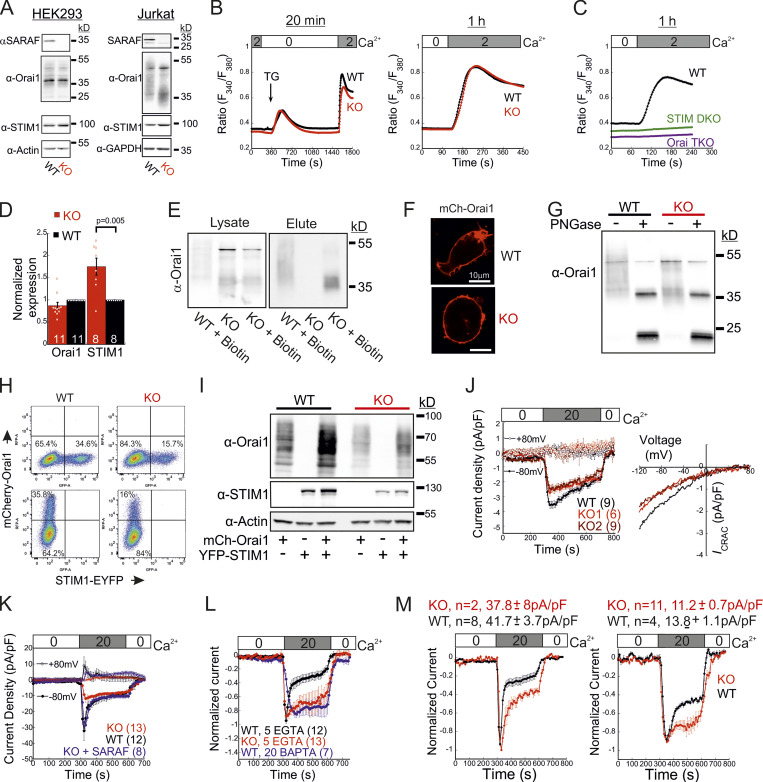
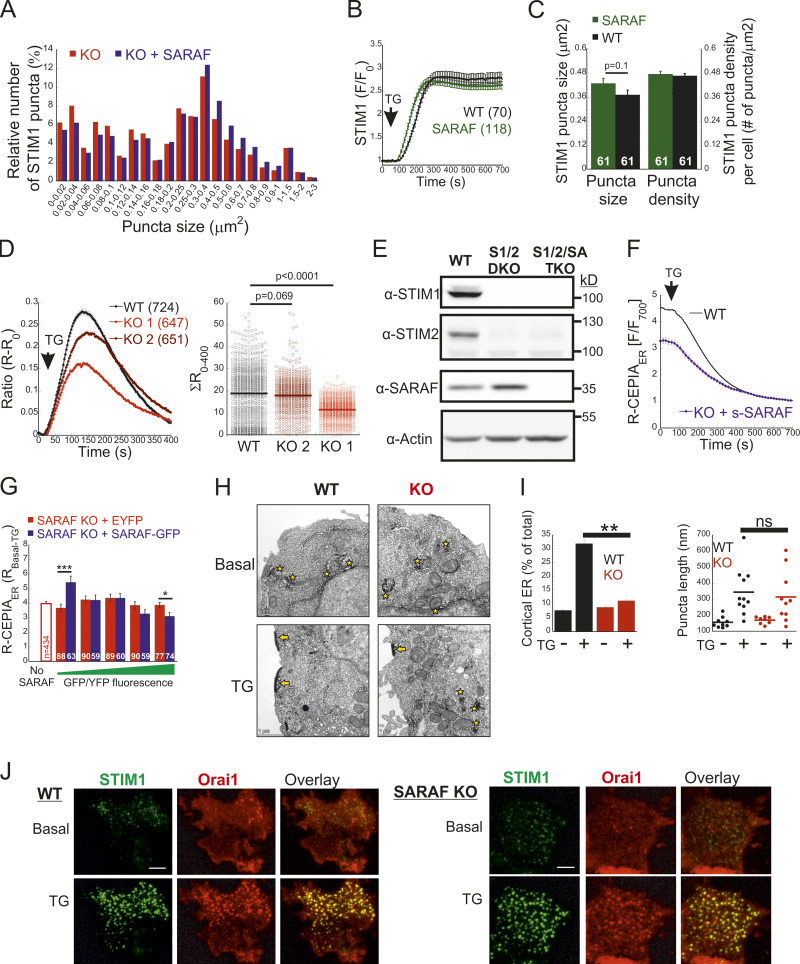
To study the role of SARAF in CRAC channel activity regulation, we used CRISPR/Cas9-mediated gene editing to ablate SARAF expression in HEK293 and Jurkat T cells (Fig. 1 A and Fig. S1 A). Surprisingly, following ER Ca2+ depletion by thapsigargin (TG) treatment for 10 min and induction of SOCE by adding back of extracellular Ca2+, SARAF knockout (KO) cells had a reduced rate and magnitude of SOCE compared with WT cells (Fig. 1 B). This difference in SOCE between WT and SARAF KO cells was diminished in a time-dependent manner when cells were treated with TG for 20 min or for 1 h before SOCE induction (Fig. S1 B). Ca2+ entry under these conditions was mediated exclusively by SOCE as adding back of extracellular Ca2+ after 1-h incubation with TG did not elicit Ca2+ entry in cells with expression KO of STIM1 and STIM2 (S1S2 double knockout [DKO]) or Orai1, Orai2, and Orai3 (Orai triple knockout [TKO]; Fig. S1 C). As shown in Fig. S1, attenuation of SOCE in SARAF KO cells did not result from deficiency in STIM1 or Orai1 expression or from poor membrane localization or altered post-translational modifications of Orai1 (Fig. S1, D–I). Because changes in intracellular Ca2+ can arise from alterations in different cellular factors such as membrane potential or activity of PM Ca2+ pumps, we employed patch-clamp electrophysiology to directly measure Ca2+ entry by CRAC channels. We recorded currents mediated by endogenously expressed channels in Jurkat T cells and by heterologously expressed channels in HEK293 cells under conditions that also permit assessment of SCDI. To activate CRAC channels, the intracellular pipette solution contained TG (1 µM) and the Ca2+ chelating agent EGTA (5 mM). After 5 min without extracellular Ca2+, addition of Ca2+ to the bath solution induced a typical ICRAC in both WT and SARAF KO Jurkat T cells (Fig. 1 C and Fig. S1 J). WT cells had larger peak current densities than SARAF KO cells, and similar results were observed in two different SARAF KO Jurkat T cell clones (Fig. 1 C and Fig. S1 J). Because EGTA captures incoming Ca2+ slowly, Ca2+ flux through CRAC channels will produce a temporary rise in free intracellular Ca2+ levels and evoke SCDI. Fig. 1 C shows that SCDI was reduced in SARAF KO cells and that changes in SCDI between WT and SARAF KO cells were independent of current magnitude (Fig. 1 D). Next, we assessed CRAC channel activity in HEK293 cells heterologously expressing EYFP-STIM1 and mCherry-Orai1. Similar to findings in Jurkat T cells, the averaged peak current densities measured in SARAF KO cells were smaller than in WT HEK293 cells (Fig. 1 E). We also observed that, following perfusion with 20 mM Ca2+, although all cells initially exhibited currents with a typical inward rectification, some cells occasionally developed small transient outward currents (Fig. S1 K). This phenomenon was absent in Jurkat cells (Fig. S1 J) and was eliminated when HEK293 cells were superfused with 2 mM Ca2+ (Fig. 4 F) and therefore was not further investigated in this study. WT cells coexpressing STIM1 and Orai1 showed significant SCDI that was attenuated by replacement of intracellular EGTA (5 mM) with a faster Ca2+ chelator [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid [BAPTA] 20 mM; Fig. S1 L]. Consistent with a key role for SARAF in SCDI, inactivation was significantly reduced in SARAF KO cells (Fig. 1 E and Fig. S1, K–M). Importantly, reconstitution of SARAF expression to SARAF KO cells restored both current inactivation and peak current density to levels observed in WT cells (Fig. 1 E), confirming that the observed changes in CRAC channel activity were brought about by the lack of SARAF expression. Taken together, these findings suggest that SARAF plays a dual regulatory role in CRAC channel function, contributing initially to channel activation and successively to channel inactivation.
Figure 1.
SARAF regulates current magnitude and SCDI of CRAC channels. (A) WB analysis of SARAF, GAPDH, and actin expression in WT and SARAF KO HEK293 and Jurkat cells, as indicated. (B) Mean ± SEM intracellular Ca2+ responses (left panels) and quantification of SOCE (right panels) in WT (black) or SARAF KO HEK293 (red) or SARAF KO Jurkat (light/dark red) cell lines. n = 152–295 cells and n = 22–48 cells in each group of Jurkat and HEK293 cells, respectively. (C) Time course of mean ± SEM current densities recorded for WT or SARAF KO Jurkat cells following superfusion with the indicated calcium concentration (in mM) at a holding potential of −120 mV (left). (D) Left: The degree of current activation (peak current densities at 330 s) plotted as a function of the degree of current inactivation (percentage current remaining at 700 s) for individual WT and SARAF KO Jurkat T cells. Right: Bar graphs show the average values from all cells. CD, current density. (E) Left: Time course of mean ± SEM currents measured in WT or SARAF KO HEK293 cells coexpressing EYFP-STIM1 and mCherry-Orai1 without or together with SARAF (blue). Right: Bar graphs show the mean ± SEM from all cells. All numbers in parentheses represent the number of cells. P values were calculated with one-way ANOVA using Tukey’s post hoc test in B and E and using a two-tailed Student’s t test in D. *, P < 0.05; **, P < 0.01.
Figure S1.
Expression and function of SARAF, STIM1, and Orai1 in SARAF KO cells. (A) WB analysis of total cell lysate using antibodies against SARAF, Orai1, STIM1, actin, or GAPDH in HEK293 (left) or Jurkat cells (right). Note that this analysis rules out the possibility that lower SOCE resulted from lower expression of STIM1 or Orai1 in either HEK293 or Jurkat T cells. (B) Mean ± SEM intracellular Ca2+ responses of SOCE in WT or SARAF KO HEK293 cells following incubation with TG for 20 min (WT, n = 85; KO, n = 83) or for 1 h (WT, n = 338; KO, n = 366). (C) Mean ± SEM intracellular Ca2+ responses of SOCE in WT, S1/S2 DKO, or Orai1/Orai2/Orai3 TKO HEK293 cells following incubation with TG for 1 h. (D) Quantification of WB analyses using antibodies against STIM1 or Orai1 in Jurkat WT and SARAF KO cells. Results show the average fraction (± SEM) of STIM1 or Orai1 expression levels in SARAF KO cells compared with WT cells. P value was calculated using a two-tailed Student’s t test. (E) Jurkat cells were biotinylated, and surface proteins were isolated with streptavidin resin, eluted, and assessed by WB analysis using α-Orai1 antibody. (F) Fluorescence images of WT and SARAF KO HEK293 cells expressing mCherry-Orai1 showing their PM localization. Scale bar, 10 μm. (G) WB analysis of lysates from WT or SARAF KO Jurkat cells before and after treatment with endoglycosidase (PNGase F), as indicated. Note that this analysis shows a decrease in glycosylation of Orai1 in SARAF KO cells compared with WT cells, which is expected to enhance rather than decrease SOCE and hence unlikely to underlie the observed decrease in CRAC channel activity in SARAF KO cells. (H and I) WT or SARAF HEK293 cells were transfected with mCherry-Orai1 and EYFP-STIM1 constructs. 30 h after transfection, cells were divided into two fractions. One fraction of cells was analyzed by flow cytometry (H) and the other by WB analysis using antibodies against STIM1, Orai1, and actin (I). Note that the reduction in STIM1 or Orai1 expression in SARAF KO cells compared with WT cells shown in I (∼50% as assessed by band density) corresponds to a similar reduction in the percentage of cells ectopically expressing STIM1 or Orai1 shown in H. (J) Left: Time course of the averaged current density (±SEM) recorded from individual WT (black) or two different clones of SARAF KO Jurkat cells (light or dark red) at −80 mV (filled circles) and +80 mV (open circles). Right: Current–voltage plots of currents from SARAF KO or WT cells, as indicated. (K) Time course of the averaged current densities (±SEM) recorded from individual WT (black) or SARAF KO HEK293 cells expressing mCherry-Orai1 together with EFYP-STIM1 (red) at −80 mV (filled circles) and +80 mV (open circles). Blue traces correspond to currents recorded from SARAF KO cells expressing mCherry-Orai1, EFYP-STIM1, and SARAF. (L) Time course of the average normalized currents (±SEM) recorded from WT (black or blue) or SARAF KO (red) HEK293 cells expressing mCherry-Orai1 and EFYP-STIM1. Pipette solution contained 1 µM TG with either 5 mM EGTA or 20 mM BAPTA, as indicated. (M) Currents from WT (black) or SARAF KO (red) HEK293 cells coexpressing EYFP-STIM1 and mCherry-Orai1 were pooled into two groups based on current size (number of cells and average current density in each group are shown on top), and the time course of the average normalized currents is shown. Note that SCDI is reduced in SARAF KO cells independently of current size. All numbers in parentheses or within bars for quantification represent the number of cells.
Figure 4.
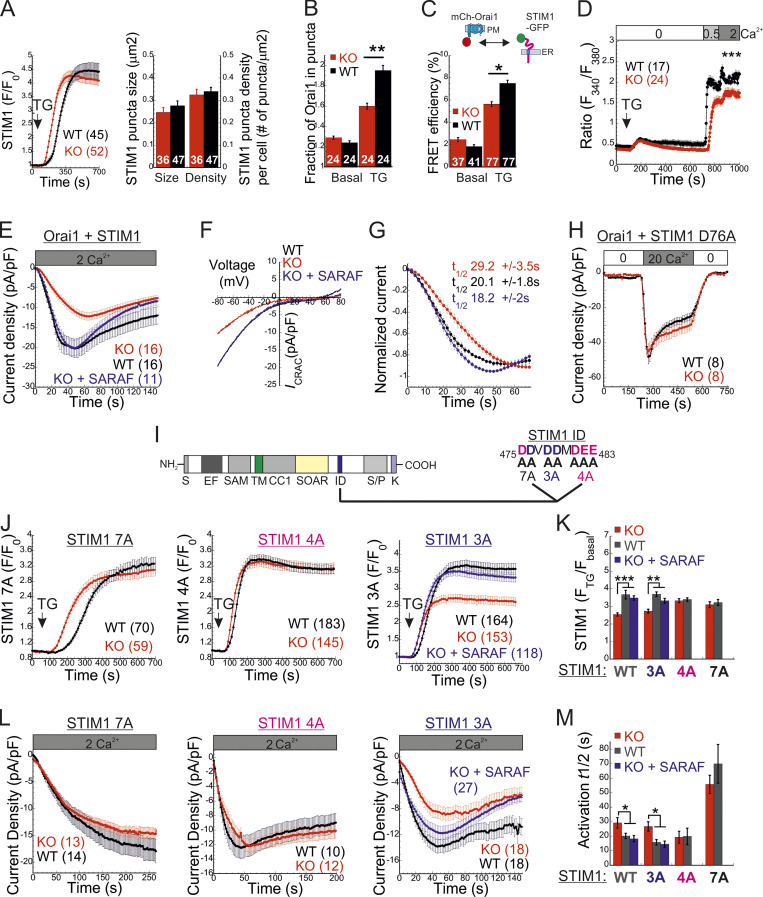
SARAF facilitates CRAC channel activation by removing an activation constraint imposed by the STIM1 ID. (A) Left: Average time course (± SEM) of changes in EYFP-STIM1 fluorescence in cells expressing EYFP-STIM1 and Orai1 at the PM following ER Ca2+ depletion by TG (1 µM). Right: Quantification of the mean ± SEM size and number of EYFP-STIM1 puncta per cell in WT and SARAF KO cells following treatment with TG for 10 min. (B) Quantification of the mean ± SEM fraction of mCherry-Orai1 fluorescence in EYFP-STIM1 puncta before and after TG treatment. (C) Quantification of the mean ± SEM FRET efficiency measured in cells coexpressing STIM1-EGFP and mCherry-Orai1 before and after TG treatment. (D) Averaged intracellular Ca2+ responses (±SEM) of SOCE in WT or SARAF KO HEK293 cells coexpressing STIM1 and Orai1. (E and F) Time course of mean ± SEM current densities (E) and plots of current–voltage (F) of whole-cell currents in HEK293 cells expressing STIM1 and Orai1 and infused with 50 µM IP3 under 2 mM external calcium are shown for the indicated number of WT (black), SARAF KO (red), or SARAF KO rescue (blue) cells. (G) Time course of mean ± SEM normalized current development shown for data in E. The mean ± SEM half-maximal activation times (t1/2) are indicated. (H) Time course of mean ± SEM currents measured in WT or SARAF KO HEK293 cells coexpressing STIM1-EYFP D76A and mCherry-Orai1. (I) Illustration of different domains of STIM1 (left), the STIM1 ID sequence, and the location of the 7A (black), 4A (magenta), and 3A (blue) mutations (right). (J) Time course of average changes (± SEM) in EYFP-STIM1 fluorescence at the PM following ER Ca2+ depletion by TG (1 µM). (K) Quantification of the averaged (± SEM) STIM1 translocation to ER–PM junctions after TG treatment in cells expressing the indicated EYFP-STIM1 mutant with or without SARAF. (L) Time course of averaged currents (± SEM) recorded from WT or SARAF KO cells coexpressing EYFP-Orai1 together with 7A, 4A, or 3A STIM1-mCherry mutants. (M) Quantification of the mean ± SEM half times for current development in cells expressing Orai1 together with the indicated EYFP-STIM1 mutant with or without SARAF. All numbers in parentheses or within bars for quantification represent the number of cells. P values were calculated with one-way ANOVA using Tukey’s post hoc test in K and M and using a two-tailed Student’s t test in B–D. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
SARAF contributes to activation of STIM1 and STIM2
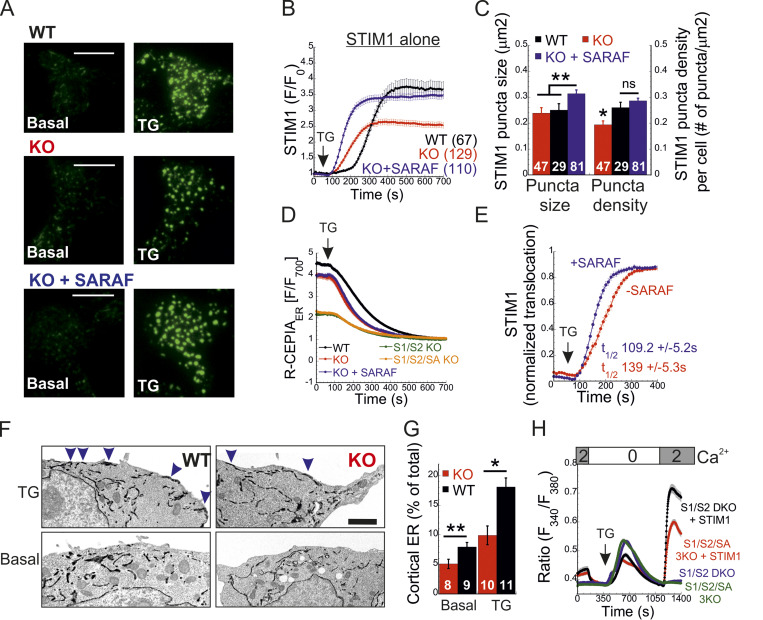
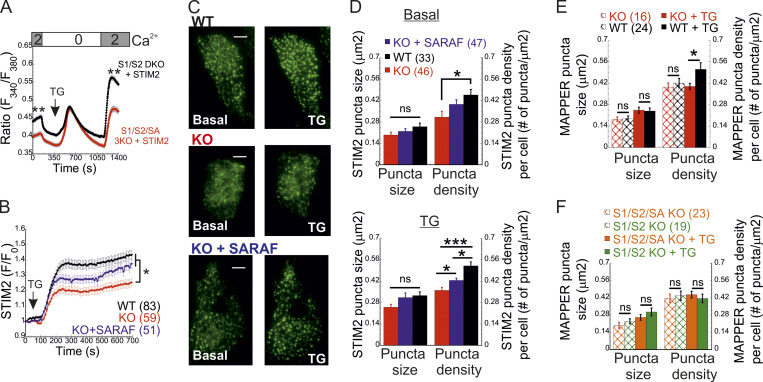
To elucidate how SARAF regulates ICRAC activation, we investigated whether translocation of STIM1 or STIM2 to ER-PM regions and whether the interaction between STIM1 and Orai1 in these regions are altered in SARAF KO cells. We initially measured the translocation of STIM1 to ER-PM sites by using total internal reflection fluorescence microscopy (TIRFM) in cells expressing EYFP-STIM1. Following passive depletion of ER Ca2+ with TG, the onset of STIM1 translocation in SARAF KO or SARAF rescue cells started earlier than in WT cells; however, the final overall degree of STIM1 recruitment at ER–PM junctions was significantly reduced in SARAF KO cells as compared with SARAF rescue cells or WT cells (Fig. 2, A and B). Analysis of the size and density of individual STIM1 puncta in these cells revealed that although the average puncta size was similar in WT and SARAF KO cells, it was significantly increased in SARAF rescue cells, and the number of puncta was significantly smaller in SARAF KO cells than in WT or SARAF rescue cells (Figs. 2 C and S2 A). Although the kinetics of STIM1 translocation to ER–PM junctions were slightly increased in WT cells overexpressing SARAF as compared with control cells, the magnitude of response and the size or number of STIM1 puncta were similar, suggesting that the effect of SARAF on activation of STIM1 is maximal already at the endogenous levels of the protein (Fig. S2, B and C). We hypothesized that the early onset of STIM1 translocation in SARAF KO cells may result from reduction in the levels of resting ER Ca2+, which shortens the time to reach the activation threshold of STIM1. Comparison of the time course of ER Ca2+ release following application of TG in WT, SARAF KO, and SARAF rescue cells revealed a significant decrease in luminal Ca2+ in SARAF KO cells as compared with WT cells, confirming that the early onset of STIM1 translocation in these cells is caused by reduced luminal Ca2+ levels (Fig. 2 D). The effect of deletion of SARAF on ER Ca2+ release was also observed in both lines of SARAF KO Jurkat T cells as compared with WT cells; however, this effect reached statistical significance only in one of the two lines of cells (Fig. S2 D).
Figure 2.
SARAF facilitates translocation of STIM1 to ER–PM junctions and promotes Ca2+ entry by STIM1. (A and B) Representative TIRFM images (A) and plot of the mean ± SEM time course (B) of changes in EYFP-STIM1 fluorescence (F) in WT or SARAF KO cells expressing EYFP-STIM1 alone or together with SARAF (blue trace) at the PM following ER Ca2+ depletion by TG (1 µM). Scale bar, 10 µm. (C) Quantification of the mean size and number (±SEM) of EYFP-STIM1 puncta per cell in WT, SARAF KO, and SARAF rescue cells following treatment with TG for 10 min. (D) ER Ca2+ was measured by tracking the fluorescence of R-CEPIA (Kd = 565 ± 58 µM) in WT (n = 545), SARAF KO (n = 243), S1/S2 DKO (n = 167), or S1/S2/SA TKO (n = 166) cells or in SARAF KO cells reexpressing SARAF (n = 478) following treatment with TG (1 µM). Plot shows the time course of the mean ± SEM normalized R-CEPIA fluorescence. (E) The mean ± SEM normalized change of EYFP-STIM1 fluorescence at the PM in SARAF KO and rescue cells following treatment with TG (1 µM). (F) Representative EM images of WT and KO cells expressing ER-targeted peroxidase (ER-HRP) and STIM1 before and after TG application. Scale bar, 2 µm. (G) Quantification of cortical ER (i.e., ER–PM junctions; blue arrows) before and after TG treatment. (H) The mean ± SEM intracellular Ca2+ responses of SOCE are shown for untransfected S1/S2 DKO (n = 130) or S1/S2/SA TKO (n = 150) cells and for S1/S2 DKO (n = 127) or S1/S2/SA TKO (n = 188) cells expressing EYFP-STIM1. All numbers in parentheses or within bars for quantification represent the number of cells. P values were calculated with one-way ANOVA using Tukey’s post hoc test in C and using a two-tailed Student’s t test in G. *, P < 0.05; **, P < 0.01.
Figure S2.
Effects of SARAF on translocation of STIM1 and STIM2 to ER–PM junctions and on ER Ca2+ release. (A) Size distribution of STIM1 puncta in SARAF KO cells and SARAF rescue cells. (B) Average time course (±SEM) of changes in EYFP-STIM1 fluorescence (F) at the PM in WT HEK293 cells expressing EYFP-STIM1 alone or together with SARAF following ER Ca2+ depletion by TG (1 µM). (C) Quantification of the size and number of EYFP-STIM1 puncta per cell in control or SARAF-overexpressing WT cells following treatment with TG for 10 min. (D) Time course of averaged change (±SEM) in cytosolic Ca2+ (left) and quantification of area under the curve from individual WT or SARAF KO Jurkat T cells (right) following treatment with TG (1 µM). (E) WB analysis of total cell lysate using antibodies against SARAF, STIM2, STIM1, or actin in WT, S1/S2 DKO, and S1/S2/SA TKO HEK293 cells. (F) Time course of average change (±SEM) in ER Ca2+ levels following treatment with TG in SARAF KO cells expressing a short splice isoform of SARAF (n = 94). Trace of WT cells from Fig. 2 D is presented for comparison. (G) ER Ca2+ levels and GFP or YFP expression were measured in SARAF KO cells transfected with SARAF-GFP or YFP (control). Expression units that cover the entire fluorescent protein fluorescence range were created by grouping the indicated number of cells distributed within similar fluorescent values (SARAF-GFP, n = 315; control-YFP, n = 434). The averaged resting ER Ca2+ levels (±SEM) are plotted as a function of expression groups. (H) Representative images of STIM1-APEX2 staining in WT and SARAF KO cells before and after TG treatment. Arrows and stars indicate clustering of STIM1 at ER–PM junction (arrows) or at intracellular structures (stars). Scale bar, 1 μm. (I) Quantification (mean ± SEM) of distribution (left) and size (right) of STIM1-APEX2 at ER–PM junctions (cortical ER, arrows, right panel) before and after treatment with TG (1 µM). (J) Representative images of WT and KO cells expressing EYFP-STIM1 and mCherry-Orai1 before and after TG application. Scale bar, 5 µm. P values were calculated using a two-tailed Student’s t test in C, G, and I and using one-way ANOVA with Tukey’s post hoc test in D. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All numbers in parentheses or within bars for quantification represent the number of cells.
To test if the changes in ER Ca2+ levels were related to changes in SOCE, we measured ER Ca2+ levels following deletion of both STIM1 and STIM2 together or without SARAF. Resting levels of ER Ca2+ were similarly reduced in cells with deletion of both STIM1 and STIM2 (S1/S2 DKO) and in cells with triple deletion of STIM1, STIM2, and SARAF (S1/S2/SA TKO), indicating that the reduced ER Ca2+ levels in SARAF KO cells resulted from a change in steady-state ER Ca2+ refilling via SOCE (Fig. 2 D and Fig. S2 E). However, reexpression of either the full-length or short splice variants of SARAF in SARAF KO cells (Fig. 2 D and Fig. S2 F) did not recover ER Ca2+ to WT levels, suggesting that resetting of ER Ca2+ back to WT levels requires the native levels of SARAF proteins or may take place over a longer time period during which the activity or expression of other components of the ER Ca2+ homeostatic mechanisms are remodeled. To assess the relationship between levels of SARAF expression and ER Ca2+, we expressed either EYFP (control) or SARAF-GFP in SARAF KO cells and measured ER Ca2+ levels as a function of protein expression, as estimated by protein fluorescence. Consistent with the effect of reexpression of untagged SARAF, the average levels of ER Ca2+ in the whole population of control and SARAF-GFP cells were similar. However, a gradual increase in expression of SARAF-GFP but not EYFP was correlated with a gradual decrease in levels of ER Ca2+ in cells, suggesting that moderate changes in expression levels of SARAF are able to fine-tune resting ER Ca2+ levels (Fig. S2 G). The similar levels of resting ER Ca2+ in SARAF KO and SARAF rescue cells allowed comparison of the rate of STIM1 translocation to ER-PM sites after depletion of ER Ca2+ with TG. This analysis indicated that reexpression of SARAF accelerates STIM1 translocation (Fig. 2 E). EM studies provided additional evidence for the contribution of SARAF to translocation of STIM1 to ER–PM junctions. Formation of ER–PM junctions was assessed before and after stimulation with TG in cells coexpressing ER-targeted HRP together with STIM1 or in cells expressing STIM1 tagged with the engineered peroxidase apurinic/apyrimidinic endodeoxyribonuclease 2 (STIM1-APEX2). Consistent with results from TIRFM analysis, although the average length of ER–PM junctions after ER Ca2+ depletion was similar between WT and SARAF KO cells (344.2 ± 44.1 nm and 314.2 ± 50 nm in WT and SARAF KO cells, respectively), SARAF KO cells had fewer ER–PM junctions containing STIM1 than did WT cells (Fig. 2, F and G; and Fig. S2, H and I).
Because STIM1 and STIM2 form heterodimers that comigrate to ER–PM contact sites, we further tested whether regulation of STIM1 activation by SARAF involved STIM2. We measured SOCE in S1/S2 DKO and S1/S2/SA TKO cells following reexpression of either STIM1 (Fig. 2 H) or STIM2 (Fig. 3 A). In either case, Ca2+ entry decreased in S1/S2/SA TKO cells as compared with S1/S2 DKO cells, indicating that SARAF regulates the activation of each STIM isoform in an independent manner. Consistent with these findings, translocation of STIM2 to ER–PM junctions after ER Ca2+ depletion was reduced in SARAF KO cells as compared with WT cells, and reexpression of SARAF in SARAF KO cells significantly recovered this deficiency (Fig. 3, B and C). Deletion of SARAF affected the density but not the size of STIM2 puncta (Fig. 3, C and D). Under resting conditions, although the average size of STIM2 puncta was similar in either WT, SARAF KO, or SARAF rescue cells, the density of STIM2 puncta was reduced in SARAF KO cells as compared with WT cells, and reexpression of SARAF in SARAF KO cells partially recovered this reduction (Fig. 3, C and D). After incubation with TG for 10 min, both the density and the size of STIM2 increased in all cell types; however, WT and SARAF rescue cells had a significantly larger puncta density than SARAF KO cells (Fig. 3, C and D). Analysis of ER–PM junctions using membrane-attached peripheral ER (MAPPER; Chang et al., 2013) provided additional evidence for a contribution of SARAF to remodeling of ER–PM junctions after ER Ca2+ depletion. Although deletion of SARAF had no effect on the size of MAPPER puncta before and after ER Ca2+ depletion, it caused a significant reduction in the density of MAPPER puncta after ER Ca2+ depletion (Fig. 3 E). Demonstrating that SARAF affects ER–PM contact sites via STIM proteins, deletion of both STIM proteins eliminated the effect of SARAF on the density of MAPPER puncta (Fig. 3 F).
Figure 3.
SARAF promotes Ca2+ entry by STIM2 and facilitates translocation of STIM2 to ER–PM junctions. (A) The averaged intracellular Ca2+ responses of SOCE are shown for S1/S2 DKO (n = 143) or S1/S2/SA TKO (n = 113) cells expressing EYFP-STIM2. (B and C) Plot of the average time course (B) and representative TIRFM images (C) of changes in EYFP-STIM1 fluorescence (F) in WT or SARAF KO cells expressing EYFP-STIM2 alone or together with SARAF (blue trace), as indicated, at the PM following ER Ca2+ depletion by TG (1 µM). Scale bar, 5 µm. (D) Quantification of the size and number of EYFP-STIM2 puncta per cell in WT, SARAF KO, and SARAF rescue cells at rest (basal, upper panel) and after treatment with TG for 10 min (TG, lower panel). (E and F) Quantification of the size and number of GFP-MAPPER puncta per cell in (E) WT or SARAF KO cells and in (F) S1/S2 DKO or S1/S2/SA TKO cells at rest and after treatment with TG for 10 min, as indicated. Mean values with SEM are shown in all panels (except C), and all numbers in parentheses represent the number of cells. P values were calculated with one-way ANOVA using Tukey’s post hoc test in B and D and using a two-tailed Student’s t test in E and F. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Taken together, the above results indicate that SARAF contributes to clustering of both STIM proteins at ER–PM contact sites. We next turned to study the contribution of SARAF to the translocation of STIM1 to ER-PM sites in cells coexpressing Orai1. In both WT and SARAF KO cells coexpressing mCherry-Orai1 and EYFP-STIM1, the degree of STIM1 translocation and the size and density of STIM1 puncta were similar (Fig. 4 A). However, analysis of Orai1 relocation into STIM1 puncta showed that less Orai1 was recruited to STIM1 puncta in SARAF KO cells than in WT cells (Fig. 4 B and Fig. S2 J). Additional analysis of the interaction between STIM1 and Orai1 was done by using Förster resonance energy transfer (FRET). FRET was determined by analyzing the recovery of donor fluorescence after acceptor photobleaching in cells coexpressing (donor) EGFP-tagged STIM1 (STIM1-GFP) and (acceptor) Orai1 (mCherry-Orai1). At resting conditions, both WT and SARAF KO cells had similar low levels of FRET (∼2%); however, following ER Ca2+ depletion with TG for 10 min, FRET increased to 7.5 ± 0.3% in WT cells but only to 5.6 ± 0.2% in SARAF KO cells, indicating that, under these conditions SARAF facilitates the interaction of STIM1 with Orai1 (Fig. 4 C). Consistent with reduced coupling between STIM1 and Orai1 in SARAF KO cells, analysis of SOCE in cells expressing EYFP-STIM1 and mCherry-Orai1 showed reduced Ca2+ entry in SARAF KO cells as compared with WT cells (Fig. 4 D). Taken together, the above results show that, after ER Ca2+ depletion, SARAF promotes the activation of STIM1 and coupling with Orai1.
SARAF cooperates with the STIM1 ID to regulate CRAC channel function
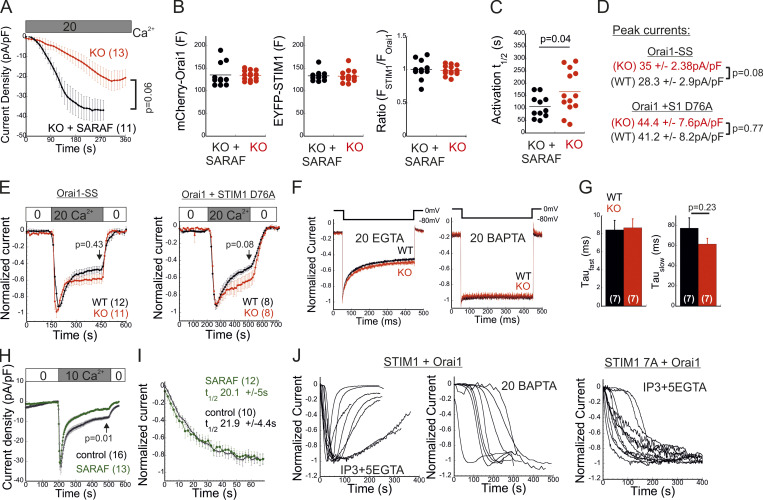
Having shown that SARAF affects both activation of STIM1 and coupling to Orai1, we next asked whether SARAF facilitates the kinetics of Orai1 channel activation by STIM1. Because SARAF KO cells and SARAF rescue cells exhibit similar resting ER Ca2+ levels (Fig. 2 D), we first analyzed the time course of development of ICRACs induced by passive ER Ca2+ depletion. Dialysis of the cytosol with a high concentration of BAPTA triggered currents with faster activation kinetics in SARAF rescue cells than in SARAF KO cells (Fig. S3, A–C). To compare the kinetics of current activation between WT and SARAF KO cells, we employed a different protocol in which high levels of inositol 1,4,5-trisphosphate (IP3; 50 µM) together with modest levels of EGTA (5 mM) are infused into the cellular cytoplasm via the patch pipette. The rapid depletion of ER Ca2+ by the IP3 receptor pathway shortens the time required for reaching the Ca2+ threshold for STIM1 activation and therefore allows comparison of channel activation kinetics in WT and SARAF KO cells in spite of the difference in resting ER Ca2+ levels in these cells. Consistent with the contribution of SARAF to CRAC channel activation, the average rate of current development was significantly slower in SARAF KO cells, and smaller current densities developed than those in WT cells (Fig. 4, E–G). Further confirming that the differences in current development were caused by SARAF, activation kinetics similar to those of WT cells were observed when SARAF was reintroduced in SARAF KO cells (Fig. 4, E–G). Using experimental protocols similar to those in Figs. 1 E and 4 E, we also tested whether increased expression of SARAF affects SCDI and the kinetics of current activation, respectively, in WT cells coexpressing STIM1 and Orai1. Consistent with earlier reports (Palty et al., 2012), increased expression of SARAF increased SCDI but not current size as compared with control cells (Fig. S3 H). Likewise, the kinetics of current activation were similar between SARAF-overexpressing and control cells (Fig. S3 I). Taken together with the small effect of overexpression of SARAF on recruitment of STIM1 to ER–PM junctions (Fig. S2, B and C), these findings indicate that the endogenous expression levels of SARAF in HEK293 cells are sufficient for promoting CRAC channel activation. To further examine how SARAF regulates CRAC channel activation, we next sought to look into whether SARAF affects channel activity when coupling between STIM1 and Orai1 is at steady state. To this end, we used Orai1 with a tandem pair of genetically fused SOAR-like fragments (Orai1-SS) as well as the constitutively active STIM1 D76A mutant. Strong channel activity was recorded in both SARAF KO and WT cells expressing Orai1-SS as well as in cells coexpressing mCherry-Orai1 and STIM1 D76A (Fig. 4 H; and Fig. S3, D and E). Interestingly, we also observed that SARAF KO cells expressing STIM1 D76A displayed a weaker tendency for current inactivation than WT cells and that current density of Orai1-SS was larger in SARAF KO cells than in WT cells; however, neither of these effects reached statistical significance (Fig. S3, D and E). Together with the observation that prolonged store depletion, which enables more time for STIM1 puncta formation and Orai1 channel trapping, restores SOCE in SARAF KO cells (Fig. S1 B), these results show that SARAF plays a rate-controlling, but not essential, role during channel activation.
Figure S3.
Effects of SARAF and the STIM1 ID on FCDI and SCDI. (A) Time course of mean ± SEM whole-cell currents in HEK293 cells expressing EYFP-STIM1 and mCherry-Orai1 and infused with 20 mM BAPTA under 20 mM external calcium are shown for the indicated number of SARAF KO (red) or SARAF KO rescue (black) cells. (B) Expression levels of STIM1 and Orai1 are shown for each cell. F, fluorescence. (C) Values of t1/2 of current activation are shown for each cell. (D and E) Average current densities (D) and time course of the mean ± SEM normalized currents (E) recorded from WT or SARAF KO HEK293 cells expressing Orai1-SS-GFP or coexpressing mCherry-Orai1 and STIM1-EYFP D76A, as indicated. Pipette solution contained 5 mM EGTA. (F) Representative traces showing currents during hyperpolarizing steps (shown on top) from 0 to −80 mV in WT (black) or SARAF KO (red) cells expressing STIM1 and Orai1. (G) The fast and slow τ values of current decay were extracted by double-exponential fitting, and the summary of averaged values is shown. Note that SARAF does not contribute to FCDI. (H) Time course of average whole-cell currents (± SEM) recorded from WT HEK293 cells coexpressing STIM1 and Orai1 with or without SARAF, as indicated. Pipette solution contained 5 mM EGTA and 1 µM TG. (I) The averaged time course (±SEM) of normalized currents recorded from WT HEK293 cells coexpressing Orai1 and STIM1 with or without SARAF, as indicated. Intracellular pipette solution contained 5 mM EGTA and 50 µM IP3. (J) Time course of normalized currents recorded from individual HEK293 cells expressing Orai1 and WT STIM1 (left) or STIM1 7A (right). Intracellular pipette solution contained either 5 mM EGTA and 50 µM IP3 or 20 mM BAPTA, as indicated. Note that although currents induced by WT STIM1 undergo slow inactivation, those induced by STIM1 7A do not. All numbers in parentheses or within bars for quantification represent the number of cells.
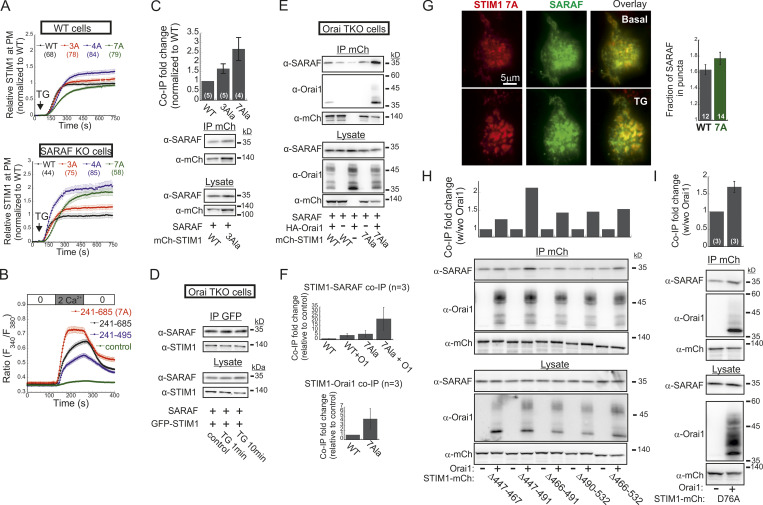
The STIM1 ID is a putative Ca2+ binding domain (Mullins et al., 2009) that consists of a stretch of conserved acidic residues (Fig. 4 I) shown to regulate FCDI (Derler et al., 2009; Mullins et al., 2009; Lee et al., 2009) and to stabilize the resting conformation of STIM1 (Lee et al., 2019). Because the interaction between SARAF and STIM1 is sensitive to deletion mutations that include or are proximal to the STIM1 ID (Jha et al., 2013), we asked whether SARAF cooperates functionally with this region in STIM1 to execute bidirectional regulation of channel function. Substitution of seven acidic residues in the STIM1 ID with alanines (STIM1 7A) eliminated both types of channel regulation by SARAF. The STIM1 7A mutant did not display preclustering before ER Ca2+ depletion, and, importantly, treatment with TG caused a similar degree of STIM1 7A translocation close to the PM in both WT and SARAF KO cells, albeit with significantly slower kinetics than WT STIM1 (Fig. 4, J and K; and Fig. S4 A). When coexpressed with Orai1, STIM1 7A displayed store-operated activation and mediated IP3-induced currents with similar magnitude and kinetics in both WT and SARAF KO cells (Fig. 4, L and M). Consistent with the slow translocation to ER–PM junctions of STIM1 7A, the kinetics of Orai1 current activation by this mutant were also significantly slower than that of WT STIM1 (Fig. 4 M). Strikingly, although a low concentration of EGTA was used for intracellular Ca2+ chelation, currents mediated by STIM1 7A did not inactivate over time. Because currents mediated by WT STIM1 slowly inactivated under identical conditions but not in the presence of BAPTA (Fig. S3 J), these findings demonstrate that STIM1 7A does not undergo SCDI. The above findings show that CRAC channel regulation by SARAF depends on the STIM1 ID and that neutralization of the charged residues in the STIM1 ID stabilizes an STIM1 conformation that is no longer sensitive to this regulation. Considering the key role of the STIM1 ID in FCDI, these findings raised the possibility that SARAF may play a role in both types of CDI. However, analysis of FCDI showed similar fast inactivation in both SARAF KO and WT cells (Fig. S3, F and G), indicating that SARAF is not required for FCDI. Because the STIM1 ID residues D476, D478, and D479 are critical for FCDI but D475, D481, E482, and E483 are not (Mullins and Lewis, 2016), we hypothesized that only the latter group of residues is required for channel regulation by SARAF. Analyses of STIM1 translocation to ER–PM junctions and development of ICRAC confirmed this hypothesis. Compared with WT cells, SARAF KO cells expressing STIM1 with a triple alanine substitution in D476, D478, and D479 (STIM1 3A) exhibited reduced accumulation at ER–PM junctions (Fig. 4, J and K), and when STIM1 3A was coexpressed with Orai1, IP3-induced currents had slower development and reduced density (Fig. 4, L and M). Reexpression of SARAF in SARAF KO cells rescued the magnitude of STIM1 3A translocation to ER–PM junctions and the kinetics of Orai1 current activation by this mutant to WT levels. Formation of ER–PM junctions and the Orai1 channel activation kinetics and peak current densities were similar in both WT and SARAF KO cells expressing an STIM1 4A mutant harboring a quadruple alanine substitution in D475, D481, E482, and E483 (Fig. 4, J–M; and Fig. S4 A). In aggregate, the above findings show that a group of acidic residues within the STIM1 ID region is critical for the bidirectional regulation of CRAC channel activity by SARAF.
Figure S4.
Effects of mutations in the STIM1 ID, CTID, and EF-hand Ca2+ binding domain of STIM1 on interaction with SARAF or Orai1 and on translocation to ER–PM junctions. (A) The indicated EYFP-STIM1 WT or ID mutants were expressed either in WT (upper panel) or SARAF KO cells (lower panel), and the time course of EYFP fluorescence changes at the PM was recorded following ER Ca2+ depletion by TG (1 µM) for each group of constructs on the same day. The time course of fluorescence change for each construct was normalized to the maximal fluorescence response recorded in control cells (expressing WT STIM1), and the mean ± SEM values of responses for each construct are plotted. (B) Averaged intracellular Ca2+ responses (± SEM) recorded in WT cells expressing HA-Orai1 alone (control, n = 223) or together with mCherry-CC1-SOAR-ID-GFP (aa 241–495, n = 163) or the WT (n = 158) or 7A mutant (n = 69) forms of the mCherry-S1CT-GFP construct (aa 241–685) following addition and washout of Ca2+ (2 mM) in the extracellular solution. (C) WB analysis of cell lysate and protein material following IP of mCherry (mCh) in cells expressing SARAF together with WT or 3A mutant STIM1. Data from Fig. 5 L for STIM1 7A are shown for comparison. (D) WB analysis of cell lysate and protein material following IP of GFP in Orai1 TKO cells expressing SARAF together with EYFP-STIM1 before and after treatment with TG (1 µM) for 1 or 10 min, as indicated. (E) WB analysis of cell lysate and protein material following IP of mCherry in Orai TKO cells expressing SARAF together with either WT or 7A mutant STIM1-mCherry with or without Orai1-HA, as indicated. (F) Summary of the relative amount of SARAF and Orai1 in IP material from Orai TKO cells expressing SARAF, HA-Orai1, and WT or 7A STIM1-mCherry. Values are normalized to those of WT STIM1-mCherry expressing cells ± SEM. (G) Representative TIRF images of a cell expressing STIM1(7A)-mCherry and SARAF-GFP and HA-Orai1 before and after application of TG (400 nM). Right: Quantification of SARAF-GFP fluorescence in STIM1-mCherry or STIM1(7A)-mCherry puncta after TG treatment. Scale bar, 4 µm. (H and I) WB analysis of cell lysate or immunoprecipitated material (IP with anti-mCherry) prepared from HEK293 SARAF KO cells expressing SARAF together with the indicated STIM1-mCherry ΔCTID constructs (H) or with STIM1-mCherry D76A (I) alone or together with HA-Orai1. Top panels show the relative change of SARAF in the IP material following expression of HA-Orai1. All numbers in parentheses or within bars for quantification represent the number of cells.
SARAF promotes a conformational change in the C-terminus of STIM1
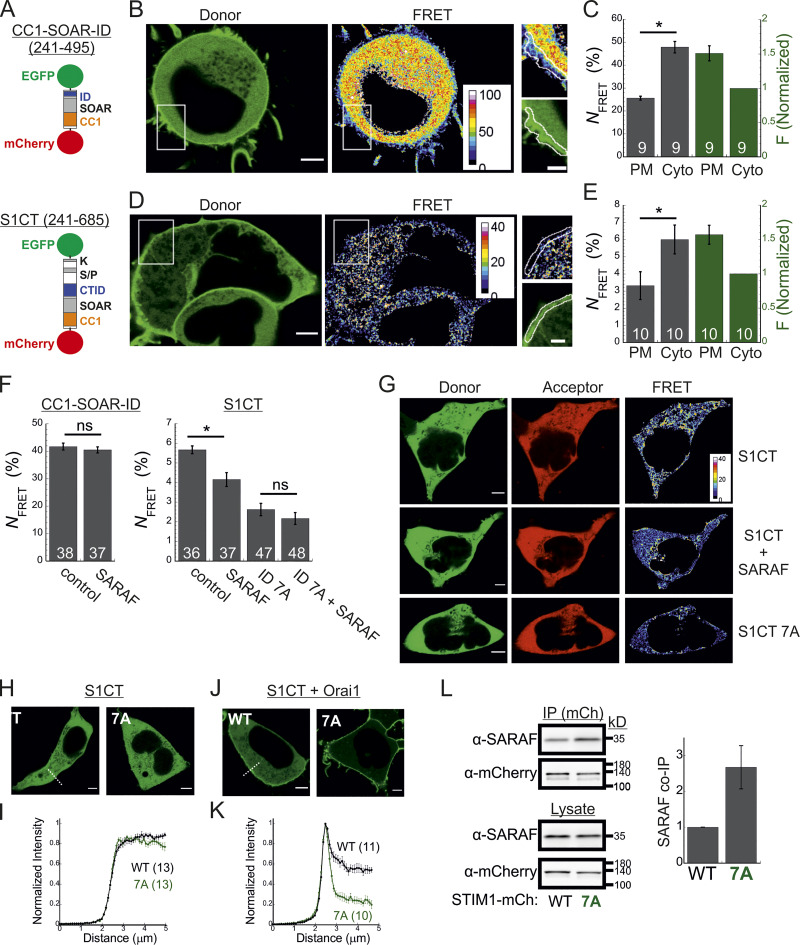
The STIM1 ID has recently been shown to stabilize the resting state of STIM1 via intramolecular interaction with the CC1 region (Lee et al., 2019). Hence, an important question is whether SARAF contributes to displacement of the ID from CC1. To address this question, we created two STIM1 conformational sensors by fusing mCherry and EGFP to the N- and C-termini, respectively, of the entire cytosol-facing C-terminal region of STIM1 (aa 241–685; mCherry-S1CT-GFP) or to a shorter fragment containing the CC1-SOAR-ID regions (aa 241–495; mCherry-CC1-SOAR-ID-GFP; Fig. 5 A). As previously established by Muik et al. (2011), evaluation of intramolecular FRET between the two fluorescent proteins allows detection of conformational changes in the C-terminus of STIM1. Consistent with earlier work (Muik et al., 2011), when either FRET sensor was coexpressed with Orai1, some of the probe was localized to the cell membrane, and FRET in these regions was significantly reduced compared with FRET in the cytosol (Fig. 5, B–E). When expressed alone, each sensor was distributed throughout the cytoplasm and exhibited either robust (∼45% for mCherry-CC1-SOAR-ID-GFP) or weak (∼6% for mCherry-S1CT-GFP) FRET. These results indicate that following coupling to Orai1, rearrangements in the C-terminus of STIM1 that are associated with the exposure of SOAR cause a decrease in FRET in both short (CC1-SOAR-ID) and full-length (S1CT) sensors. We next examined whether SARAF affects FRET of either sensor. Expression of SARAF had no effect on FRET in mCherry-CC1-SOAR-ID-GFP (Fig. 5 F). In contrast, expression of SARAF caused a reduction in FRET in mCherry-S1CT-GFP, suggesting that SARAF affects the arrangement of a region found C-terminally to the STIM1 ID (Fig. 5 F). Consistent with a key role for the STIM1 ID in setting the resting conformational state, an mCherry-S1CT-GFP sensor harboring the 7A mutation exhibited increased binding to Orai1 channels (Fig. 5, H–K) and activation of Orai1 channels (Fig. S4 B) and had reduced FRET compared with the WT mCherry-S1CT-GFP (Fig. 5, F and G). Notably, expression of SARAF did not induce a further decrease in FRET of mCherry-S1CT(7A)-GFP, suggesting that SARAF binding to STIM1 and neutralization of the ID acidic residues may stabilize a similar STIM1 conformation (Fig. 5 F). If the latter is correct, we expected an increase in binding between SARAF and STIM1 7A compared with WT STIM1. Supporting this idea, coimmunoprecipitation (co-IP) experiments showed that the interaction between SARAF and STIM1 7A was increased by ∼2.5-fold compared with WT STIM1 (Fig. 5 L). Interaction between SARAF and STIM1 3A was also increased compared with WT STIM1, but only by ∼65% (Fig. S4 C). Taken together, these results show that SARAF promotes a conformational change in the C-terminus of STIM1 that depends on the STIM1 ID and involves rearrangement of a region located C-terminally to the STIM1 ID.
Figure 5.
SARAF promotes a conformational change in the C-terminus of STIM1. (A) Illustration of the two conformational STIM1 sensors used in this work. (B–E) Analysis of conformational changes in the STIM1 CT using FRET sensors. (B and D) Representative images of donor and FRET signals in a cell expressing mCherry-CC1-SOAR-ID-GFP (B) or mCherry-S1CT-GFP (D) together with Orai1. White squares show the regions enlarged on the right. Scale bar, 7 µm (full images) and 2 µm (insets). (C and E) The average FRET and overall sensor fluorescence measured at the PM or cytosol (Cyto). Sensor fluorescence at the PM is normalized to fluorescence recorded in the cytosol. (F) Bar graph shows the averaged FRET for the indicated sensors expressed in SARAF KO cells alone (control) or together with reexpression of SARAF. (G) Representative images of donor, acceptor, and FRET images from SARAF KO cells expressing mCherry-S1CT-GFP, mCherry-S1CT(7A)-GFP, or mCherry-S1CT-GFP together with SARAF. (H–K) Representative fluorescence images (H and J) and summary of normalized intensity values (I and K) across the PM and cytosolic regions (dashed lines indicate representative regions used for analysis) of cells expressing mCherry-S1CT-GFP or mCherry-S1CT(7A)-GFP expressed without (H and I) or together with Orai1 (J and K). Scale bar, 4 µm. (L) WB analysis of cell lysate or immunoprecipitated material (IP using anti-mCherry) prepared from HEK293 SARAF KO cells expressing SARAF without STIM1-mCherry or STIM1(7A)-mCherry. Summary of the relative amount of SARAF in IP material is shown (right) with values normalized to SARAF in total lysate (n = 4). Mean values with SEM are shown in panels C, E, F, I, K, and L, and all numbers in parentheses or within bars for quantification represent the number of cells. P values were calculated with a two-tailed Student’s t test in C, E, and F. *, P < 0.05.
SARAF binds to STIM1-Orai1 channel complex independently of SCDI
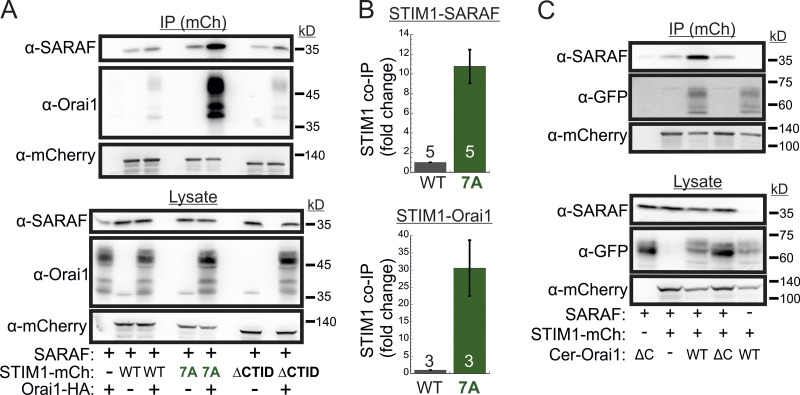
We and others had previously shown that SARAF and STIM1 interact under resting conditions and that this interaction increases when STIM1 binds to Orai1 after ER Ca2+ depletion (Maléth et al., 2014; Palty et al., 2012). Western blot (WB) analysis of STIM1 co-IP material extracted from Orai TKO cells coexpressing EYFP-STIM1 together with SARAF showed similar levels of SARAF in the co-IP material (Fig. S4 D), indicating that interaction between STIM1 and SARAF under either replete or depleted ER Ca2+ occurs independently of Orai1. Consistent with earlier reports, this interaction was significantly increased upon reexpression of Orai1 (Fig. S4, E and F). Because the interaction between SARAF and STIM1 also increases in the presence of the SCDI-insensitive STIM1 7A mutant independently of Orai (Fig. S4, E and F), we asked if SARAF interacts with this STIM1 mutant when it is in complex with Orai1. Similar to WT STIM1, following ER Ca2+ depletion, STIM1 7A accumulated at ER-PM regions and recruited SARAF into punctate structures, suggesting that SARAF interacts with the STIM1 7A mutant when in complex with Orai1 (Fig. S4 G). WB analysis of STIM1 co-IP material from cells expressing HA-Orai1, SARAF, and STIM1-mCherry after ER Ca2+ depletion demonstrated a strong increase in the interaction of STIM1 7A with either SARAF (∼10-fold increase) or Orai1 (∼30-fold increase) as compared with WT STIM1 (Fig. 6, A and B). This interaction was diminished in cells expressing a deletion mutant of Orai1 that lacks the C-terminal STIM1 binding region (Orai1 ΔC) as compared with cells expressing WT Orai1 (Fig. 6 C), indicating that neutralization of the ID acidic residues strongly stabilizes an active SARAF-STIM1-Orai1 channel complex. The strong interaction between STIM1 7A and SARAF is in apparent contradiction with findings from a recent study which showed that a number of SCDI-resistant STIM1 mutants, including several CTID deletion mutants and the EF-hand domain D76A mutant, display poor binding to SARAF (Jha et al., 2013). We therefore extended the STIM1 co-IP analysis to investigate the interaction of SARAF with different STIM1 ΔCTID mutants and with STIM1 D76A in the absence or presence of Orai1. As can be seen in Fig. S4, H and I, the interaction between SARAF and STIM1 ΔCTID or STIM1 D76A increases in the presence of Orai1, indicating that SARAF interacts differentially with STIM1, depending on whether it is in complex with Orai1. Because STIM1 7A, STIM1 ΔCTID, and STIM1 D76A mutants all exhibit reduced or no SCDI, these results confirm that SARAF interacts with the STIM1-Orai1 complex independently of SCDI.
Figure 6.
SARAF binds to the STIM1-Orai1 channel complex independently of SCDI. (A) WB analysis of cell lysate and IP material following IP of mCherry (mCh) in cells expressing the indicated combinations of SARAF, HA-Orai1, and WT, 7A, or ΔCTID (Δ466–530) STIM1-mCherry constructs. (B) Summary of the relative amount of SARAF and Orai1 in IP material from cells expressing SARAF, HA-Orai1, and WT or 7A STIM1-mCherry. Values are normalized to those of WT STIM1-mCherry–expressing cells ± SEM, and numbers within bars for quantification represent the number of repeats. (C) WB analysis of cell lysate and IP material following IP of mCherry in cells expressing the indicated combinations of SARAF, STIM1 (7A)-mCherry, and the indicated WT or ΔC (Δ273–301) Cerulean-Orai1 constructs.
SARAF contributes to NFAT-mediated transcription in T cells
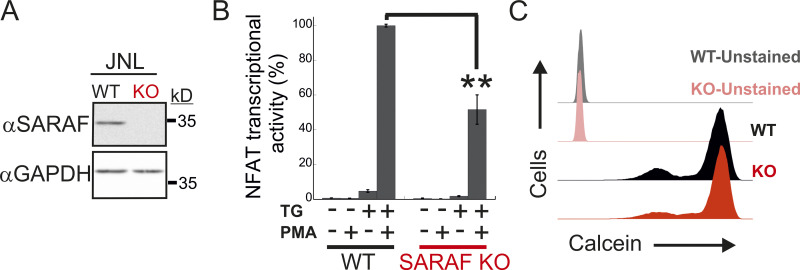
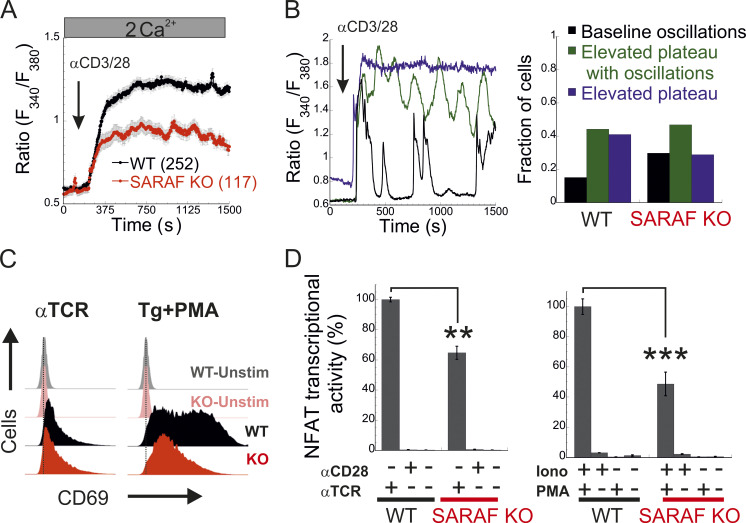
Having shown that SARAF regulates CRAC channel function in a bidirectional manner, we sought to assess the physiological significance of this activity during cellular signaling in T lymphocytes. Stimulation of the TCR by cognate antigen leads to the release of Ca2+ from the ER and consequently to ER Ca2+ depletion and activation of CRAC channels. Ca2+ flux through CRAC channels elevates intracellular Ca2+ levels and stimulates the Ca2+-calcineurin-NFAT signaling pathway, which in turn regulates the expression of a large number of T cell activation–related genes (Hogan et al., 2003; Macián et al., 2002). To determine the contribution of SARAF to Ca2+-dependent transcription in T cells, we analyzed TCR-stimulated Ca2+ signaling and gene expression, both in Jurkat cells and in a subline of these cells that contains an NFAT-inducible luciferase reporter gene (Jurkat NFAT luciferase [JNL] cells). To mimic TCR activation by antigen-presenting cells, Jurkat T cells were stimulated with αCD3/αCD28-coated beads, and intracellular Ca2+ responses were monitored both in WT cells and in SARAF KO cells that we generated (Fig. S5 A). Here, in the absence of SARAF, the average intracellular Ca2+ response to stimulus over 20 min was significantly reduced compared with that in WT cells (Fig. 7 A). At the single-cell level, TCR activation generated primarily three different intracellular Ca2+ responses (Fig. 7 B): repetitive Ca2+ responses that returned to starting baseline after each peak (baseline oscillations), repetitive Ca2+ responses with elevated baseline levels (elevated plateau oscillations), and responses that generated a sustained plateau of intracellular Ca2+ (elevated plateau). The initial response event to TCR stimulation was similar in both WT (0.59 ± 0.017 fura-2 ratio units) and SARAF KO cells (0.54 ± 0.021 fura-2 ratio units), and a similar proportion of cells in each cell type responded with Ca2+ oscillations with an elevated plateau (44.0% for WT cells and 44.4% for SARAF KO cells). Conversely, the proportion of cells with baseline Ca2+ oscillations was about twofold larger in SARAF KO cells (28.2%) than in WT cells (15.1%), whereas the proportion of cells exhibiting the elevated plateau type of response was smaller in SARAF KO cells (27.4%) than in WT cells (40.9%). These results suggest that the stimulatory effect of SARAF on CRAC channel activity plays a dominant role over its inhibitory action under these conditions. We next evaluated transcriptional responses either following TCR stimulation or when bypassing it by application of TG or ionomycin together with PMA, an activator of protein kinase C. TCR stimulation triggered a weaker NFAT transcription response in SARAF KO JNL cells than in WT JNL cells and up-regulated the expression of the T cell early activation marker CD69 in both WT and SARAF KO Jurkat cells (Fig. 7, C and D). When cells were stimulated with TG and PMA, SARAF KO Jurkat T cells up-regulated the expression of CD69, albeit to a lesser extent than WT cells (Fig. 7 C). SARAF KO JNL cells also exhibited a reduction in NFAT transcriptional activity compared with WT JNL cells when cells were stimulated with the combination of either PMA and ionomycin (Fig. 7 D) or PMA and TG (Fig. S5 B). The observed reduction in Ca2+-dependent gene expression in SARAF KO cells was not caused by differential cellular toxicity, because both WT and SARAF KO cells exhibited similar cellular viability following stimulation with TG and PMA (Fig. S5 C). In the aggregate, the above findings indicate that SARAF contributes to T cell activation responses by promoting TCR-mediated signaling via the Ca2+-calcineurin-NFAT pathway.
Figure S5.
WT and SARAF KO Jurkat cells have similar viability. (A) WB analysis of total cell lysates using antibodies against SARAF or GAPDH in WT or SARAF KO Jurkat cells harboring an NFAT-inducible luciferase reporter gene (JNL cells). (B) JNL cells were stimulated with the indicated combination of TG and PMA, and luciferase activity was used to determine NFAT-regulated gene expression 6 h after treatment. Bar graph shows the normalized mean ± SEM values obtained from three to six repetitions. **, P < 0.01. (C) Flow cytometry analysis of calcein staining in WT or SARAF KO Jurkat cells 16 h after treatment with PMA and TG.
Figure 7.
SARAF regulates gene expression by NFAT in T cells. (A) Mean ± SEM intracellular Ca2+ response recorded from WT or SARAF KO Jurkat T cells following stimulation by nanobeads conjugated to α-CD3 and α-CD28 at the indicated time point (WT, n = 252; SARAF KO, n = 117). (B) Left: Representative traces of individual cells showing the time course of the three types of intracellular Ca2+ responses to TCR stimulation: baseline oscillations (black), oscillations with elevated plateau (green), and elevated plateau without oscillations (blue). Right: Bar graph shows the relative distribution of each type of Ca2+ response in WT or SARAF KO cells. (C) Histograms from flow cytometry experiments show the distribution of cells as a measure of α-CD69-FITC fluorescence intensity with or without TCR or TG/PMA (at 1 µM/30 nM, respectively) stimulation, as indicated. (D) JNL cells were stimulated with the indicated combination of α-CD28 and α-CD3 (α-TCR) antibodies (left) or ionomycin (2 µM) and PMA (right), and luciferase activity was used to determine NFAT-regulated gene expression. Bar graph shows the normalized mean ± SEM values obtained from three to six repetitions. P values were calculated using a two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001.
Discussion
The functional contribution of SARAF to cellular Ca2+ homeostasis and CRAC channel SCDI has been widely discussed in the literature, where an increase in SARAF expression was shown to reduce the levels of resting intracellular Ca2+ and promote SCDI, whereas reduction in SARAF expression was shown to increase the levels of resting intracellular Ca2+ and reduce SCDI (Maléth et al., 2014; Jha et al., 2013; Lopez et al., 2019; Palty et al., 2012; Albarran et al., 2016a). These observations, together with evidence for an interaction between SARAF and STIM1 (Jha et al., 2013; Palty et al., 2012; Albarran et al., 2016a; Maléth et al., 2014) or SOAR (Jha et al., 2013), led to a model in which SARAF regulates CRAC channel activity through inhibitory binding to SOAR of STIM1. However, a fundamental limitation of this basic model is that it fails to explain the increase in SARAF–STIM1 interaction that occurs during coupling of STIM1 to Orai1 and before induction of SCDI (Palty et al., 2012; Maléth et al., 2014; Jha et al., 2013). The present work resolves this discrepancy and reveals that recruitment of SARAF to the CRAC channel complex contributes to an initial excitatory interaction between STIM1 and Orai1. Prior to this study, we and others employed experimental protocols designed specifically for dissecting current inactivation and consequently did not meticulously examine the contribution of SARAF to the activation of ICRACs. Indeed, although we had previously found a small decrease in the magnitude of ICRACs following RNAi-mediated knockdown of SARAF expression in Jurkat T cells (Palty et al., 2012), that difference did not reach statistical significance, and additional studies in HEK293 cells did not report a change in current densities when expression of endogenous SARAF was reduced (Jha et al., 2013; Lopez et al., 2019; Maléth et al., 2014). Hence, although abolishment of SARAF expression in HEK293 cells attenuates both channel activation and inactivation (Figs. 1 and 2), low amounts of SARAF (estimated at ∼30% of control levels; Palty et al., 2012; Jha et al., 2013) are fully sufficient for channel activation but not for inactivation, whereas further increase to or above the endogenous expression levels of SARAF only augments channel inactivation (Fig. 1 E; and Fig. S3, H and I). We do not yet fully understand the underlying mechanism behind this phenomenon; however, several explanations are possible. One possibility is that there are different stoichiometric recruitments for facilitation of CRAC channel activation and for SCDI by SARAF. Because endogenous SARAF levels are quite high in HEK293 cells, as evidenced by the ability of the native protein to enforce strong SCDI when STIM1 and Orai1 are overexpressed (Jha et al., 2013; Palty et al., 2012; Maléth et al., 2014), it is plausible that the reduction of SARAF expression in these cells with siRNAs generates a ratio of SARAF to STIM1 that supports CRAC channel activation but not SCDI. This scenario is consistent with the previously observed increase in SOCE and ER Ca2+ levels after partial reduction of SARAF expression with siRNA (Palty et al., 2012) and with the opposing effects on ER Ca2+ levels reported herein following reexpression of either low or high levels of SARAF (Fig. S2 G). An alternative scenario is that SARAF uses reversible oligomerization to switch between positive and negative regulatory modes. In this scenario, reduction of SARAF expression shifts the dimer/monomer distribution of SARAF toward monomers that are deficient in promoting SCDI but effective in facilitating CRAC channel activation. Consistent with this possibility, Kimberlin et al. (2019) recently showed that cells coexpressing STIM1 and Orai1 together with a monomeric mutant of SARAF produce larger currents than cells coexpressing WT SARAF. Further studies using SARAF mutants trapped in a monomeric or dimeric configuration are required to resolve this issue.
The role of SARAF in CRAC channel activation described herein is consistent with recent studies which show that SARAF contributes to Ca2+ entry in human endothelial and murine hybridoma cell lines (Albarran et al., 2016a; Galeano-Otero et al., 2021). It is noteworthy, however, that earlier work suggested that SARAF exerts this effect through direct interaction with Orai1 after receptor-evoked ER Ca2+ depletion (Albarran et al., 2016a). Although the present study does not rule out direct interaction between SARAF and Orai1, it strongly indicates that SARAF contributes to channel activation through an STIM1-dependent mechanism. Importantly, this finding sheds light on a hitherto elusive step during STIM1-Orai1 coupling activation in which SOAR is liberated from the CC1 lock but remains transiently suppressed by an inhibitory region located C-terminally to SOAR. This inhibitory region was initially mapped to residues 448–530, the CTID (Jha et al., 2013); however, more recent work showed that residues 470–491, which correspond to the STIM1 ID, constitute the minimal region within the CTID that mediates this effect (Lee et al., 2019). The involvement of the STIM1 ID in maintaining STIM1 in a nonactive conformation is further supported herein by the observation that alanine replacement of acidic residues within the STIM1 ID causes rearrangement in the C-terminus of STIM1 and enhances its ability to bind and activate Orai1 channels (Figs. 5 and S4 B). The transition from the resting to the activated conformation of STIM1 therefore involves the release of two sequential intramolecular brakes on SOAR, the first imposed by the CC1 region and the second by the STIM1 ID. The release of both brakes appears to be regulated by accessory proteins. Following ER Ca2+ depletion, STIMATE (TMEM110) was shown to bind directly to the CC1 region to facilitate the unlocking of SOAR from CC1 and accumulation of STIM1 at ER–PM junctions (Jing et al., 2015). Several lines of evidence derived from this study suggest that SARAF facilitates the release of the STIM1 ID brake. First, SARAF promotes the interaction of STIM1 with the PM (Fig. 2, A–C, F, and G) and with Orai1 (Fig. 4, B–G), and these effects are sensitive to mutation in the STIM1 ID (Fig. 4, J–M). Second, FRET analysis using the mCherry-S1CT-GFP conformational sensor shows that SARAF promotes a conformational change in a region that overlaps with the STIM1 ID and that this effect is also sensitive to mutation in the STIM1 ID (Fig. 5). Third, a consistent finding from co-IP analysis in the present work (Fig. 6 and Fig. S4, H and I) and from FRET analyses reported in earlier studies (Palty et al., 2012; Maléth et al., 2014; Jha et al., 2013) is that STIM1–SARAF interaction increases during coupling to Orai1 and therefore correlates with unlocking of the STIM1 ID from SOAR. Because Orai1 promotes binding of SARAF to STIM1 D76A and STIM1 7A (Fig. 6 and Fig. S4, E, F, H, and I) that exhibit reduced or no SCDI, we also conclude that initial interaction between SARAF, STIM1, and Orai1 occurs independently of SCDI.
Currently, we do not know how the rise in intracellular Ca2+ switches an initial excitatory interaction between SARAF and STIM1 to a subsequent inhibitory one. Nonetheless, the finding that the STIM1 ID is essential for both types of CRAC channel regulation by SARAF provides a first clue in resolving this question. The effect of alanine replacement of acidic residues in the STIM1 ID on slow inactivation (Figs. 4 L and S3 J), together with the contribution of these residues to Ca2+ binding (Mullins et al., 2009), suggests that the STIM1 ID functions as a Ca2+ sensor for SCDI. Because SCDI by SARAF occurs at ER–PM junctions enriched with phosphatidylinositol 4,5-bisphosphate (Maléth et al., 2014), and because SARAF affects clustering of STIM1 or STIM2 at ER–PM junctions, the lack of SCDI by STIM1 7A taken together with its slow translocation to ER–PM junctions (Fig. S4 A) may further indicate that this mutant has a different preference for membrane lipids than that of WT STIM1.
A notable finding of the present work is that SCDI is only reduced in SARAF KO cells but not completely abolished (Fig. 1, C–E; and Fig. S1 M). Consistent with the idea that SCDI arises from multiple mechanisms (Dagan and Palty, 2021), this finding demonstrates unequivocally that although SARAF is an important factor for SCDI, it is not its sole mediator of this activity in HEK293 or Jurkat T cells. This conclusion is also supported by a recent report which showed that ANO-8 contributes to a distinct type of SARAF-independent SCDI in HEK293 cells (Jha et al., 2019).
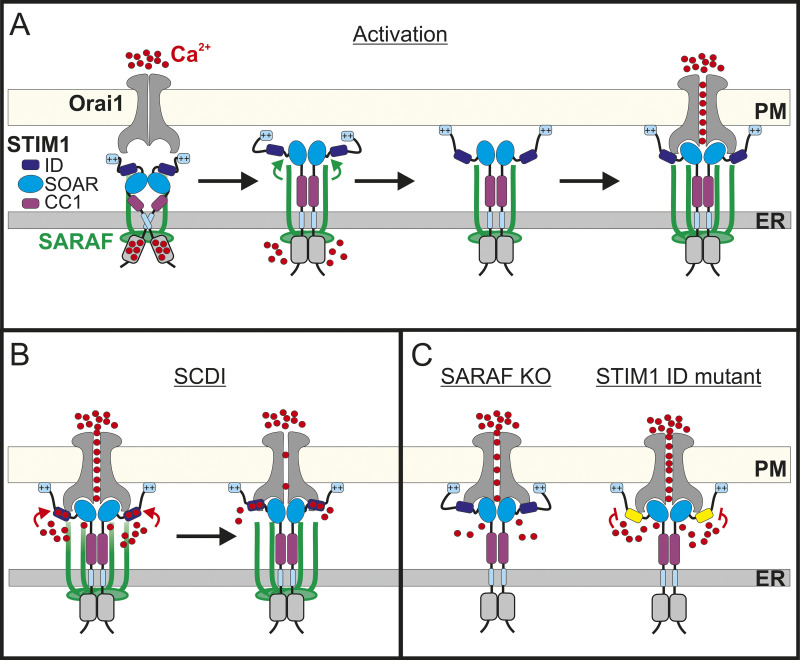
On the basis of results from both previous work and the present study, we propose the following model for CRAC channel regulation by SARAF (Fig. 8). Under resting conditions, SOAR is shielded by interactions with both CC1 and STIM1 ID. Upon Ca2+ depletion from the ER, STIM1 unfolds, the CC1 lock is removed, and STIM1 oligomerizes but SOAR remains temporarily inhibited by the STIM1 ID. Interaction of SARAF with the partially activated STIM1 facilitates a second conformational change that releases the STIM1 ID inhibition from SOAR and further extends to the lipid binding tail of STIM1. The release of the STIM1 ID brake promotes interaction of STIM1 with the PM and with the C-terminus of Orai1, resulting in channel activation at full strength. Ca2+ entry through Orai1 generates a local rise in Ca2+ levels. Electrostatic interaction of Ca2+ and negatively charged residues within the STIM1 ID (Mullins et al., 2009) triggers SCDI, and interaction of SARAF with Ca2+-bound STIM1 ID accelerates this process. Although this remains a hypothesis at this point, the model may enable a better understanding of the physiological roles of SOCE regulation by SARAF in different cell types. We previously showed that SARAF is an important factor of CRAC channel SCDI in T lymphocytes, a well-characterized in vitro model for T cell biology, but the physiological role of SARAF in these cells remained poorly defined. In the present work, we demonstrate that T lymphocytes lacking SARAF expression display impaired TCR-mediated Ca2+-calcineurin-NFAT–dependent gene expression (Fig. 7) and that this signaling deficiency arises from a reduction in Ca2+ entry through CRAC channels. Hence, the contribution of SARAF to CRAC channel activation is critical to the total Ca2+ entry in T lymphocytes. Recent studies using cells isolated from SARAF KO mice showed that although SOCE was unaffected in cardiomyocytes (Sanlialp et al., 2020), it was increased in pancreatic acinar cells (Son et al., 2019). Although it is currently unclear whether the increase in Ca2+ entry in murine acinar cells lacking SARAF expression is mediated by CRAC channels or by TRPC1 channels, which are also regulated by SARAF (Albarrán et al., 2016b) and have been shown to be strong mediators of SOCE in these cells (Hong et al., 2011), the variable effects of SARAF on SOCE regulation in T lymphocytes, cardiomyocytes, and acinar cells are consistent with the hypothesis that SARAF may affect Ca2+ entry and downstream signaling in a cell type–dependent manner.
Figure 8.
Model for CRAC channel activity regulation by SARAF. (A) Under resting conditions, STIM1 and SARAF interact, the SOAR stabilizes STIM1 dimers, and intramolecular interactions with CC1 and ID maintain SOAR in the resting conformation. Upon Ca2+ depletion from the ER, STIM1 partially unfolds; yet, the formation of ER–PM contact sites and trapping of Orai1 and channel activation are transiently inhibited by the ID. Stimulatory interaction between SARAF and STIM1 relieves the ID inhibition and promotes interaction of STIM1 with PM lipids. Recruitment of additional SARAF molecules facilitates coupling activation between STIM1 and Orai1. (B) Channel activation at full strength leads to a rise in intracellular Ca2+ close to the channel that induces negative feedback through electrostatic interactions of Ca2+ with negatively charged residues within the ID. (C) Left: Both activation and SCDI are attenuated in the absence of SARAF. Right: Mutation of Ca2+ binding residues within the STIM1 ID eliminates SCDI.
In summary, the dual regulatory effect of SARAF on CRAC channel activity presented in this work sheds light on a poorly characterized step during CRAC channel activation and provides an important framework for understanding the physiological role of SARAF in different cell types and its potential involvement in different pathophysiological conditions, including pancreatitis (Son et al., 2019), cancer (Romanuik et al., 2009; Iżykowska et al., 2017), neurodegenerative diseases (Iżykowska et al., 2017; Taha et al., 2015; Twine et al., 2011), and cardiovascular diseases (Yang et al., 2019; Dai et al., 2018; Camargo and Azuaje, 2008; Sanlialp et al., 2020; La Russa et al., 2020).
Materials and methods
Cell culture and transfection
The Jurkat and HEK293 cells were obtained from the University of California, Berkeley, Molecular and Cell Biology Tissue Culture Facility and were cultured in RPMI or MEM, respectively, as previously described (Palty et al., 2012). JNL cells (Signosis) stably expressing a firefly luciferase reporter gene under the control of the NFAT response element were a generous gift from Michael Kienzler (University of Connecticut, Storrs, CT). The STIM1/STIM2 DKO cells (Emrich et al., 2019) were a generous gift from Mohamed Trebak (University of Pittsburgh, Pittsburgh, PA). The STIM1/STIM2/SARAF TKO cells were generated using STIM DKO cells and the pX330sgSARAF plasmid as described below and were generously gifted by Eitan Reuveny (Weizmann Institute of Science, Rehovot, Israel). The Orai TKO cells (Alansary et al., 2020) were a generous gift from Barbara Niemeyer (Saarland University, Saarbrücken, Germany). Plasmid transfection of cells was performed using polyethylenimine 25K (Polysciences) or Lipofectamine 2000 (Invitrogen) according to the manufacturers’ protocols. For electrophysiological experiments, cells were plated onto an 18-mm coverglass coated with poly-L-lysine 6–8 h after plasmid transfection and 12–15 h before the start time of experiments. For FRET and intracellular Ca2+ imaging experiments, cells were plated onto an 18-mm coverglass coated with poly-L-lysine, and culture medium was replaced to wash the transfection reagent away 6–8 h after plasmid transfection. To avoid constitutive Ca2+ level elevation in cells expressing a constitutively active form of Orai1 or STIM1, cells were cultured in high-glucose, Ca2+-free DMEM supplemented with 50 μM La3+.
Generation of SARAF KO cells
The pX330sgSARAF plasmid targeting the SARAF sequence 5′-AGACTACAAAGGCGATCCCA-3′ was a generous gift from Eitan Reuveny (Weizmann Institute of Science, Rehovot, Israel). HEK293 cells were transfected, and Jurkat cells were electroporated with pX330sgSARAF and EYFP plasmids. 2 d following vector delivery, cells were sorted by flow cytometry, and EYFP-positive cells were collected. Single cells were seeded into individual wells in a 96-well plate configuration, and colonies were cultured for 10–30 d. Disruption of the SARAF gene in individual colonies was validated by sequencing the genomic loci of SARAF and by WB analysis.
Fluorescence measurements of intracellular Ca2+ ions
Cells were plated onto 18-mm coverglasses coated with poly-L-lysine. On the day of the experiment, the coverglass was mounted on an imaging chamber and washed with Ringer’s solution (140 mM NaCl, 10 mM Hepes, 10 mM glucose, 0.8 mM MgCl2, 2.8 mM KCl, and 2 mM CaCl2, pH 7.4). To load cells with fura-2-acetoxymethyl ester (Invitrogen), cells were incubated for 30 min in Ringer’s solution containing 5 μM of the Ca2+ indicator and for an additional 30 min without the dye. Live-cell imaging was performed at RT using setups built on an Olympus IX73 or IX83 microscope. To record cytosolic Ca2+ levels, fura-2-acetoxymethyl ester–loaded cells were excited by alternate illumination at 340 nm and 380 nm delivered by monochromator (Visitron), and emission was collected with a 515-nm long-pass filter. Recording of ER Ca2+ content was performed in cells expressing the ER Ca2+ reporter ER-GCaMP6-150 by illumination with 480 nm, and emission was collected with a 515-nm long-pass filter. Cells were imaged using a 20× objective, and images were acquired every 2 or 3 s with a cooled charge-coupled device camera (Retiga R3; QImaging) or with a back-illuminated scientific complementary metal-oxide semiconductor (sCMOS) camera (Prime 95B; Teledyne Photometrics). In each experiment, traces from 10 to 50 cells were recorded, and the averaged response from all cells was plotted with KaleidaGraph. All experiments were repeated at least three times. Statistical significance was determined with t test analysis for paired comparisons or using ANOVA with the Student-Newman-Keuls test for multiple comparisons. P < 0.05 was considered significant. All data are shown as mean ± SEM.
Cell surface protein biotinylation and expression analysis
Jurkat T cells were washed twice in chilled PBS solution, then incubated on ice for 30 min with freshly prepared 2 mM EZ-Link Sulfo-NHS-LC-Biotin (TS-21335; Thermo Fisher Scientific), washed twice in PBS with 100 mM glycine, followed by cell lysis as above. Cell lysates were incubated with streptavidin-conjugated agarose beads (16-126; Mercury) for 2 h on ice with agitation. The beads were washed three times in lysis buffer, and the bound proteins were eluted at 95°C for 10 min in the gel sample buffer. Proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes for WB analysis. The membranes were first blocked with 5% nonfat dry milk in TBS-T (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20), then incubated with primary antibodies for Orai1 (O8264, 1:1,000; Sigma-Aldrich), STIM1 (AB9870, 1:1,000; Mercury), SARAF (affinity-purified rabbit polyclonal antibody against the N-terminus of SARAF was kindly provided by Eitan Reuveny, Weizmann Institute of Science, Rehovot, Israel; 1:10,000), GAPDH (ab8245, 1:1,000; Abcam) or β-actin (MS-1295-P1, 1:5,000; Thermo Fisher Scientific). The SARAF antibody was produced. Membranes were incubated with the appropriate HRP-conjugated secondary antibodies (705-036-147, 115-036-003, 111-036-003; Jackson ImmunoResearch) and analyzed using an ECL detection system (Mercury or Bio-Rad Laboratories). For reblotting, the membranes were washed, incubated in stripping buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, and 0.1 M 2-mercaptoethanol) at 50°C for 30 min, then underwent the same blocking and antibody incubation protocol.
Co-IP
24 h after transfection, HEK293 cells were first washed with Ringer’s solution, incubated in Ringer’s solution containing 1 µM TG for 10 min at RT (unless indicated otherwise), then treated with chilled lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.6, 10% glycerol, 1 mM EDTA, 0.5% Triton X-100) containing protease inhibitors (Roche). Samples were incubated on ice for 15–30 min, homogenized by pipetting, then centrifuged for 20 min at 17,000 g. IP was performed using agarose-conjugated anti-RFP beads (RFP-Trap; Chromotek) by incubation in the lysis buffer for 1 h on ice with rocking. The beads were washed three times in chilled lysis buffer before elution by boiling in Laemmli sample buffer. SDS-PAGE, WB, and ECL analyses were performed as detailed above. Primary antibodies were for mCherry (ab167453, 1:1,000; Abcam), GFP (ab6658, 1:1,000; Abcam), or Orai1 (sc-377281, 1:1,000; Santa Cruz Biotechnology).
Determination of Jurkat T cell activation and cell viability
Surface expression of CD69, an early activation marker of T cells, and cell viability were analyzed by flow cytometry. For determination of CD69 expression following T cell activation, cells were incubated for 5–6 h with or without anti-TCR (C305; Merck) or with 0.5 µM TG and 30 nM PMA, as indicated. Cells were then washed twice in ice-cold PBS, stained with FITC-conjugated CD69 mAb (11-0699-42; Invitrogen) for 15 min on ice, washed, and resuspended in FACS buffer (PBS + 1% BSA, 1 mM EDTA) before analysis. For analysis of cell viability, after 16–18 h following TG/PMA stimulation, cells were incubated in 1 µM calcein AM (C3100MP; Invitrogen) for 20 min on ice, washed in FACS buffer, and subjected to flow cytometry analysis to determine cell viability.
NFAT-dependent luciferase assay
For measurements of induction of NFAT expression using luciferase as a reporter, JNL cells were stimulated for 6–18 h as indicated. Treatments were applied to 40 × 103 or 80 × 103 cells/well in 96-well plates in triplicate. Luminescence was measured using the Steady-Glo luciferase assay system (Promega). All the data were normalized against the control group.
Generation of plasmids
Plasmids encoding SARAF, SARAF-GFP, EYFP-s-SARAF, STIM1-mCherry, STIM1-EGFP, Orai1-mEGFP, FLAG-mCherry-Orai1, and HA-Orai1 were previously described (Palty et al., 2012, 2017). The Orai1 ΔC (Δ273–301) and Orai1-SS-EGFP constructs were previously described (Palty et al., 2015). The R–calcium-measuring organelle-entrapped protein indicators (CEPIA)–ER plasmid was a generous gift from Dr. Masamitsu Iino (Nihon University School of Medicine, Tokyo, Japan; plasmid 58216; Addgene); EYFP-STIM2 and EYFP-STIM1 (Huang et al., 2006) plasmids were a generous gift from Dr. Shmuel Muallem (National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD); and the GFP-MAPPER plasmid was a generous gift from Dr. Jen Liou (University of Texas Southwestern Medical Center, Dallas, TX; plasmid 117721; Addgene). A multistep procedure was used for generating the different STIM1-mCherry CTID deletion mutants and the mCherry-S1CT-GFP or mCherry-CC1-SOAR-ID-GFP FRET constructs. For STIM1-mCherry CTID deletion mutants, first SalI sites were introduced at both ends of each deletion region within the STIM1-mCherry ORF, and the corresponding region was subsequently excised by SalI digestion and religated to create the final CTID deletion construct. For FRET constructs, STIM1 ORFs corresponding to S1CT or CC1-SOAR-ID were first inserted into a p3XFLAG 7.1 plasmid containing mCherry to create mCherry-S1CT or mCherry-CC1-SOAR-ID. Then, EcoRI and AgeI sites were introduced at the flanking ends of mCherry-S1CT or mCherry-CC1-SOAR-ID, and the digested fragments were inserted instead of the corresponding sequence of Orai1 in the Orai1-mEGFP plasmid. The STIM1 3A and 4A (Mullins and Lewis, 2016) were a generous gift from Dr. Richard Lewis (Stanford University, Stanford, CA), and the STIM1 7A mutation was created with NEBuilder HiFi DNA Assembly kit (NEB-E2621S). Primer sequences used to generate the above constructs are shown in Table S1. All plasmids were sequenced at the Technion Biomedical Core Facility for verification of the modified plasmid regions.
Confocal microscopy
Midplane sections of mCherry-Orai1 or FRET constructs expressing cells were captured using an LSM 880 confocal microscope (Zeiss) equipped with a 63×/1.49 NA objective with 488- and 561-nm lasers and controlled by Zen imaging software (Zeiss). EGFP was excited with 488 nm, and emission was collected from 505 to 580 nm. mCherry was excited at 561 nm, and emission was collected from 583 to 685 nm.
TIRFM and FRET measurements
TIRFM was performed using an inverted Olympus IX83 microscope combined with a cellTIRF-4Line system (405-, 488-, 561-, 640-nm lasers) and equipped with a back-illuminated sCMOS camera (Prime 95B; Teledyne Photometrics). Fluorescence was collected using a 100×/1.49 NA objective.
For FRET measurements using recovery of donor fluorescence after acceptor photobleaching, fluorescence signals were split into GFP (525/50) and mCherry (650/70) channels through a dual-view device. Images were acquired and processed using CellSens software and exported as Microsoft Excel files. FRET efficiency was calculated as the percentage of FRET efficiency = 100 × (Dpost − Dpre)/Dpost, where Dpre and Dpost are the donor (EGFP) intensities before and after acceptor (mCherry) bleaching, respectively.
For FRET measurements using a STIM1-based conformational sensor donor, acceptor and FRET images were acquired using an LSM 880 confocal microscope, and normalized FRET images were generated by using the pixFRET plugin in Fiji.
Analysis of STIM1, STIM2, and MAPPER puncta
Identification and segmentation of individual STIM1, STIM2, or MAPPER puncta were performed with Fiji using the Weka trainable segmentation plugin that enables machine learning–based image processing (Arganda-Carreras et al., 2017). Segmentation training was performed with manual annotation of STIM1 puncta in three different TIRF images of SARAF rescue cells. The resulting classifier was then tested on six different TIRF images of WT cells, and results from this analysis were compared with results from an independent manual analysis of the same images. Each type of analysis generated a relatively similar number of puncta (575 and 612, respectively) with similar size distribution. After validation of the classifier, TIRF images from WT, KO, and SARAF rescue cells expressing the indicated fluorescently tagged proteins were analyzed together in one batch.
EM
Processing of samples for DAB reactions was based on a procedure reported by Martell et al. (2017). Transfected HEK293 cells were grown on Aclar film pretreated with poly-L-lysine, washed, and treated with TG (1 µM) for 10 min. Then the cells were fixed on ice for 1 h with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 2 mM CaCl2. Prechilled buffers and reagents were used, and the cells were kept on ice for all subsequent steps. The cells were washed with cacodylate buffer, incubated for 5 min with 20 mM glycine in the buffer, and rinsed again. A freshly prepared solution containing 0.5 mg/ml DAB and 0.03% H2O2 was added to the cells; the generation of reaction product was monitored by transmitted light microscopy and stopped by washing with buffer. The washed cells were postfixed using 2% osmium tetroxide for 30 min, washed, and incubated in 1% uranyl acetate for 1 h. Following dehydration in graded ethanol series, the Aclar films with the cells were moved to fresh wells filled with Epon 812 to start embedding. After polymerization of Epon, the Aclar film was pealed from the block face, and 75-nm transverse sections were cut using an ultramicrotome UC7 (Leica Microsystems), transferred to copper grids, and viewed using a Talos L120C transmission electron microscope. Analysis of ER–PM junctions and localization of STIM1 at these sites was performed with Fiji.
Electrophysiology
Jurkat T cells were cultured in RPMI supplemented with 10% FBS and 1% penicillin/streptomycin. Prior to recordings, cells were placed onto poly-L-lysine–coated coverglass for 5–15 min and subsequently washed with Ringer’s solution and transferred to the recording chamber (ALA Scientific).
HEK293 cells were cultured in MEM supplemented with 10% FBS and 1% penicillin/streptomycin. ∼6–8 h after transfection, cells were transferred to poly-L-lysine–coated coverglass and maintained overnight in serum containing antibiotic-free MEM culture. 24–36 h after transfection, cells were washed with Ringer’s solution and placed in a recording chamber. Membrane currents were recorded with an integrated patch-clamp amplifier (Sutter Instrument Co.) under voltage-clamp conditions using the whole-cell variation of the patch-clamp procedure. Patch pipettes were fabricated from borosilicate glass capillaries (2–7 MΩ). Signals were analogue filtered with a 5-kHz low-pass Bessel filter. Data acquisition and analysis were carried out with SutterPatch software (Sutter Instrument Co.). All data were leak corrected by subtracting the current recorded in either Ca2+-free or Ca2+-containing recording solution supplemented with 10–100 mM La3+ or by subtracting the first trace recorded at the start of whole-cell recording before activation of STIM1, as appropriate in each case. Current densities were calculated by normalizing currents measured at −80 mV (HEK293 cells) or at −120 mV (Jurkat T cells) to cell capacitance. Three recording protocols were employed. The first was employed to assess FCDI and consisted a 400-ms step to −80 mV from a holding potential of 0 mV delivered every minute. The second protocol was employed to assess the kinetics of current activation and consisted a 20-ms step to −80 mV followed by a 100-ms ramp to +80 mV delivered every 2 s. The third protocol was employed to assess SCDI and consisted of a 20-ms step to −120 mV (for Jurkat T cells) or to −80 mV (for HEK293 cells), followed by a 100-ms ramp to +80 mV and delivered every 10 s. External Ringer’s solution contained 145 mM NaCl, 2.8 mM KCl, 10 mM Hepes, and 10 mM glucose (pH 7.4 with NaOH). CaCl2 (20 mM or 2 mM) or MgCl2 (20 mM) was added to the external solution for establishing a high- or low-Ca2+ or Ca2+-free solutions, respectively. The internal solution contained 150 mM Cs aspartate, 8 mM MgCl2, 10 mM Hepes, and 2 mM MgATP (pH 7.2 with CsOH). To chelate intracellular Ca2+, either 20 mM BAPTA or 20 mM or 5 mM EGTA was included in the intracellular solution.
Statistical analysis
Comparison of two groups was performed with the standard Student’s t test, and comparison of more than two groups was carried out with ANOVA with Tukey’s post hoc test. Comparison of puncta size distributions was performed with an unpaired Wilcoxon-Mann-Whitney rank-sum test.
Online supplemental material
Fig. S1 includes information regarding the expression and function of SARAF, STIM1, and Orai1 in SARAF KO cells. Fig. S2 includes information on the effects of SARAF on the translocation of STIM1 and STIM2 to ER–PM junctions and on ER Ca2+ release. Fig. S3 describes the effects of SARAF and the STIM1 ID on FCDI and SCDI. Fig. S4 includes data regarding the effects of mutations in the STIM1 ID, CTID, and EF-hand Ca2+ binding domain of STIM1 on interaction with SARAF or Orai1 and on translocation to ER–PM junctions. Fig. S5 shows the effects of SARAF on TG- and PMA-induced NFAT transcription and on cellular viability in Jurkat cells. Table S1 shows the sequences of primers that were used to generate the different constructs reported in this study.
Supplementary Material
shows the sequences of primers that were used to generate the different constructs reported in this study.
Acknowledgments
The authors thank Dr. Ronald Udasin for his helpful comments and critical reading of the manuscript.
The work was supported by the Rappaport Family Institute for Research in Medical Sciences and the Israel Science Foundation (ISF grant 1048/18 to R. Palty).
The authors declare no competing financial interests.
Author contributions: E. Zomot and H. Achildiev: conceptualization, data curation, formal analysis, writing – original draft, review and editing; I. Dagan and R. Militsin: data curation and formal analysis; R. Palty: conceptualization, data curation, formal analysis, funding acquisition, supervision and writing – original draft, review and editing.
References
- Alansary, D., Peckys D.B., Niemeyer B.A., and de Jonge N.. 2020. Detecting single ORAI1 proteins within the plasma membrane reveals higher-order channel complexes. J. Cell Sci. 133:jcs240358. 10.1242/jcs.240358 [DOI] [PubMed] [Google Scholar]
- Albarran, L., Lopez J.J., Amor N.B., Martin-Cano F.E., Berna-Erro A., Smani T., Salido G.M., and Rosado J.A.. 2016a. Dynamic interaction of SARAF with STIM1 and Orai1 to modulate store-operated calcium entry. Sci. Rep. 6:24452. 10.1038/srep24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarrán, L., López J.J., Gómez L.J., Salido G.M., and Rosado J.A.. 2016b. SARAF modulates TRPC1, but not TRPC6, channel function in a STIM1-independent manner. Biochem. J. 473:3581–3595. 10.1042/BCJ20160348 [DOI] [PubMed] [Google Scholar]
- Ambudkar, I.S., de Souza L.B., and Ong H.L.. 2017. TRPC1, Orai1, and STIM1 in SOCE: friends in tight spaces. Cell Calcium. 63:33–39. 10.1016/j.ceca.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arganda-Carreras, I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., and Sebastian Seung H.. 2017. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 33:2424–2426. 10.1093/bioinformatics/btx180 [DOI] [PubMed] [Google Scholar]
- Camargo, A., and Azuaje F.. 2008. Identification of dilated cardiomyopathy signature genes through gene expression and network data integration. Genomics. 92:404–413. 10.1016/j.ygeno.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Chang, C.-L., Hsieh T.-S., Yang T.T., Rothberg K.G., Azizoglu D.B., Volk E., Liao J.-C., and Liou J.. 2013. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5:813–825. 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Dagan, I., and Palty R.. 2021. Regulation of store-operated Ca2+ entry by SARAF. Cells. 10:1887. 10.3390/cells10081887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, F., Zhang Y., Wang Q., Li D., Yang Y., Ma S., and Yang D.. 2018. Overexpression of SARAF ameliorates pressure overload-induced cardiac hypertrophy through suppressing STIM1-Orai1 in mice. Cell. Physiol. Biochem. 47:817–826. 10.1159/000490036 [DOI] [PubMed] [Google Scholar]
- Derler, I., Fahrner M., Muik M., Lackner B., Schindl R., Groschner K., and Romanin C.. 2009. A Ca2+ release-activated Ca2+ (CRAC) modulatory domain (CMD) within STIM1 mediates fast Ca2+-dependent inactivation of ORAI1 channels. J. Biol. Chem. 284:24933–24938. 10.1074/jbc.C109.024083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich, S.M., Yoast R.E., Xin P., Zhang X., Pathak T., Nwokonko R., Gueguinou M.F., Subedi K.P., Zhou Y., Ambudkar I.S., et al. 2019. Cross-talk between N-terminal and C-terminal domains in stromal interaction molecule 2 (STIM2) determines enhanced STIM2 sensitivity. J. Biol. Chem. 294:6318–6332. 10.1074/jbc.RA118.006801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner, M., Muik M., Schindl R., Butorac C., Stathopulos P., Zheng L., Jardin I., Ikura M., and Romanin C.. 2014. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1). J. Biol. Chem. 289:33231–33244. 10.1074/jbc.M114.610022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano-Otero, I., Del Toro R., Khatib A.-M., Rosado J.A., Ordóñez-Fernández A., and Smani T.. 2021. SARAF and Orai1 contribute to endothelial cell activation and angiogenesis. Front. Cell Dev. Biol. 9:639952. 10.3389/fcell.2021.639952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, P.G., Chen L., Nardone J., and Rao A.. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232. 10.1101/gad.1102703 [DOI] [PubMed] [Google Scholar]
- Hong, J.H., Li Q., Kim M.S., Shin D.M., Feske S., Birnbaumer L., Cheng K.T., Ambudkar I.S., and Muallem S.. 2011. Polarized but differential localization and recruitment of STIM1, Orai1 and TRPC channels in secretory cells. Traffic. 12:232–245. 10.1111/j.1600-0854.2010.01138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G.N., Zeng W., Kim J.Y., Yuan J.P., Han L., Muallem S., and Worley P.F.. 2006. STIM1 carboxyl-terminus activates native SOC, Icrac and TRPC1 channels. Nat. Cell Biol. 8:1003–1010. 10.1038/ncb1454 [DOI] [PubMed] [Google Scholar]
- Iżykowska, K., Przybylski G.K., Gand C., Braun F.C., Grabarczyk P., Kuss A.W., Olek-Hrab K., Bastidas Torres A.N., Vermeer M.H., Zoutman W.H., et al. 2017. Genetic rearrangements result in altered gene expression and novel fusion transcripts in Sézary syndrome. Oncotarget. 8:39627–39639. 10.18632/oncotarget.17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, A., Ahuja M., Maléth J., Moreno C.M., Yuan J.P., Kim M.S., and Muallem S.. 2013. The STIM1 CTID domain determines access of SARAF to SOAR to regulate Orai1 channel function. J. Cell Biol. 202:71–79. 10.1083/jcb.201301148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, A., Chung W.Y., Vachel L., Maleth J., Lake S., Zhang G., Ahuja M., and Muallem S.. 2019. Anoctamin 8 tethers endoplasmic reticulum and plasma membrane for assembly of Ca2+ signaling complexes at the ER/PM compartment. EMBO J. 38:e101452. 10.15252/embj.2018101452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, J., He L., Sun A., Quintana A., Ding Y., Ma G., Tan P., Liang X., Zheng X., Chen L., et al. 2015. Proteomic mapping of ER–PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 17:1339–1347. 10.1038/ncb3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin, C.R., Meshcheriakova A., Palty R., Raveh A., Karbat I., Reuveny E., and Minor D.L. Jr. 2019. SARAF luminal domain structure reveals a novel domain-swapped β-sandwich fold important for SOCE modulation. J. Mol. Biol. 431:2869–2883. 10.1016/j.jmb.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzeniowski, M.K., Manjarrés I.M., Varnai P., and Balla T.. 2010. Activation of STIM1-Orai1 involves an intramolecular switching mechanism. Sci. Signal. 3:ra82. 10.1126/scisignal.2001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Russa, D., Frisina M., Secondo A., Bagetta G., and Amantea D.. 2020. Modulation of cerebral store-operated calcium entry-regulatory factor (SARAF) and peripheral Orai1 following focal cerebral ischemia and preconditioning in mice. Neuroscience. 441:8–21. 10.1016/j.neuroscience.2020.06.014 [DOI] [PubMed] [Google Scholar]
- Lee, K.P., Yuan J.P., Zeng W., So I., Worley P.F., and Muallem S.. 2009. Molecular determinants of fast Ca2+-dependent inactivation and gating of the Orai channels. Proc. Natl. Acad. Sci. USA. 106:14687–14692. 10.1073/pnas.0904664106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.K., Lee M., Jeong S.J., Qin X., Lee A.R., Park H., and Park C.Y.. 2019. IDstim helps STIM1 keep inactive via intramolecular binding to the coiled-coil domain in a resting state. J. Cell Sci. 133:jcs237354. 10.1242/jcs.237354 [DOI] [PubMed] [Google Scholar]
- Liou, J., Kim M.L., Heo W.D., Jones J.T., Myers J.W., Ferrell J.E. Jr., and Meyer T.. 2005. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 15:1235–1241. 10.1016/j.cub.2005.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, E., Frischauf I., Jardin I., Derler I., Muik M., Cantonero C., Salido G.M., Smani T., Rosado J.A., and Redondo P.C.. 2019. STIM1 phosphorylation at Y316 modulates its interaction with SARAF and the activation of SOCE and ICRAC. J. Cell Sci. 132:jcs226019. 10.1242/jcs.226019 [DOI] [PubMed] [Google Scholar]
- Lunz, V., Romanin C., and Frischauf I.. 2019. STIM1 activation of Orai1. Cell Calcium. 77:29–38. 10.1016/j.ceca.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Macián, F., García-Cózar F., Im S.-H., Horton H.F., Byrne M.C., and Rao A.. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 109:719–731. 10.1016/S0092-8674(02)00767-5 [DOI] [PubMed] [Google Scholar]
- Maléth, J., Choi S., Muallem S., and Ahuja M.. 2014. Translocation between PI(4,5)P2-poor and PI(4,5)P2-rich microdomains during store depletion determines STIM1 conformation and Orai1 gating. Nat. Commun. 5:5843. 10.1038/ncomms6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell, J.D., Deerinck T.J., Lam S.S., Ellisman M.H., and Ting A.Y.. 2017. Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells. Nat. Protoc. 12:1792–1816. 10.1038/nprot.2017.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik, M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M., et al. 2011. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. EMBO J. 30:1678–1689. 10.1038/emboj.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, F.M., and Lewis R.S.. 2016. The inactivation domain of STIM1 is functionally coupled with the Orai1 pore to enable Ca2+-dependent inactivation. J. Gen. Physiol. 147:153–164. 10.1085/jgp.201511438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, F.M., Park C.Y., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. USA. 106:15495–15500. 10.1073/pnas.0906781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty, R., Raveh A., Kaminsky I., Meller R., and Reuveny E.. 2012. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 149:425–438. 10.1016/j.cell.2012.01.055 [DOI] [PubMed] [Google Scholar]
- Palty, R., Stanley C., and Isacoff E.Y.. 2015. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 25:963–980. 10.1038/cr.2015.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty, R., Fu Z., and Isacoff E.Y.. 2017. Sequential steps of CRAC channel activation. Cell Rep. 19:1929–1939. 10.1016/j.celrep.2017.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.Y., Hoover P.J., Mullins F.M., Bachhawat P., Covington E.D., Raunser S., Walz T., Garcia K.C., Dolmetsch R.E., and Lewis R.S.. 2009. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 136:876–890. 10.1016/j.cell.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney, J.W. Jr. 1986. A model for receptor-regulated calcium entry. Cell Calcium. 7:1–12. 10.1016/0143-4160(86)90026-6 [DOI] [PubMed] [Google Scholar]
- Romanuik, T.L., Ueda T., Le N., Haile S., Yong T.M.K., Thomson T., Vessella R.L., and Sadar M.D.. 2009. Novel biomarkers for prostate cancer including noncoding transcripts. Am. J. Pathol. 175:2264–2276. 10.2353/ajpath.2009.080868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlialp, A., Schumacher D., Kiper L., Varma E., Riechert E., Ho T.C., Hofmann C., Kmietczyk V., Zimmermann F., Dlugosz S., et al. 2020. Saraf-dependent activation of mTORC1 regulates cardiac growth. J. Mol. Cell. Cardiol. 141:30–42. 10.1016/j.yjmcc.2020.03.004 [DOI] [PubMed] [Google Scholar]
- Son, A., Ahuja M., Schwartz D.M., Varga A., Swaim W., Kang N., Maleth J., Shin D.M., and Muallem S.. 2019. Ca2+ influx channel inhibitor SARAF protects mice from acute pancreatitis. Gastroenterology. 157:1660–1672.e2. 10.1053/j.gastro.2019.08.042 [DOI] [PubMed] [Google Scholar]
- Stathopulos, P.B., and Ikura M.. 2010. Partial unfolding and oligomerization of stromal interaction molecules as an initiation mechanism of store operated calcium entry. Biochem. Cell Biol. 88:175–183. 10.1139/O09-125 [DOI] [PubMed] [Google Scholar]
- Stathopulos, P.B., Li G.-Y., Plevin M.J., Ames J.B., and Ikura M.. 2006. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region: An initiation mechanism for capacitive Ca2+ entry. J. Biol. Chem. 281:35855–35862. 10.1074/jbc.M608247200 [DOI] [PubMed] [Google Scholar]
- Taha, S., Aljishi M., Alsharoqi I., and Bakhiet M.. 2015. Differential upregulation of the hypothetical transmembrane protein 66 (TMEM66) in multiple sclerosis patients with potential inflammatory response. Biomed. Rep. 3:98–104. 10.3892/br.2014.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebak, M., and Kinet J.-P.. 2019. Calcium signalling in T cells. Nat. Rev. Immunol. 19:154–169. 10.1038/s41577-018-0110-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine, N.A., Janitz K., Wilkins M.R., and Janitz M.. 2011. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One. 6:e16266. 10.1371/journal.pone.0016266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Jin H., Cai X., Li S., and Shen Y.. 2012. Structural and mechanistic insights into the activation of stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. USA. 109:5657–5662. 10.1073/pnas.1118947109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Li S., Wang Q., and Yang D.. 2019. Transmembrane protein 66 attenuates neointimal hyperplasia after carotid artery injury by SOCE inactivation. Mol. Med. Rep. 20:1436–1442. 10.3892/mmr.2019.10328 [DOI] [PubMed] [Google Scholar]
- Yuan, J.P., Zeng W., Dorwart M.R., Choi Y.-J., Worley P.F., and Muallem S.. 2009. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11:337–343. 10.1038/ncb1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Srinivasan P., Razavi S., Seymour S., Meraner P., Gudlur A., Stathopulos P.B., Ikura M., Rao A., and Hogan P.G.. 2013. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 20:973–981. 10.1038/nsmb.2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the sequences of primers that were used to generate the different constructs reported in this study.