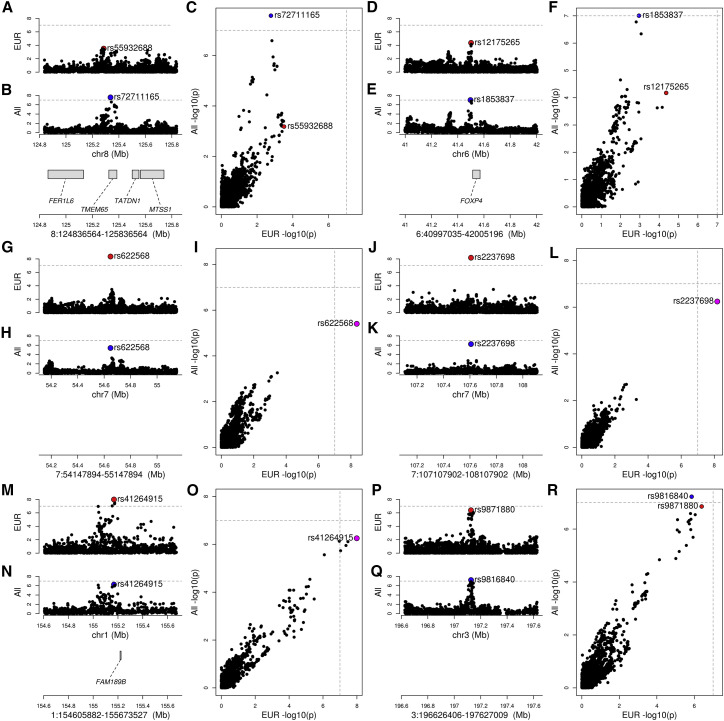
Figure 2.
Ethnicity-associated signals
Four plots are shown for each of the six loci with heterogeneous signals between European-only and multi-ethnic GWAS: (1) GWAS signal in European-only (lead variant highlighted in red); (2) GWAS signal in multi-ethnic (lead variant highlighted in blue); (3) genes with eQTLs (Table S2) overlapping GWAS locus (genes without eQTLs are not shown); and (4) scatterplot showing the -log10 (p value) calculated in the European-only (x axis) and multi-ethnic (y axis) GWAS. If the lead variant is the same between the two studies, it is colored in magenta; otherwise, the lead variant for each study is shown (red, European only; blue, multi-ethnic). For all six loci designated as heterogeneous between the ethnicities, at least two studies on the same ethnicity and phenotype (susceptibility or severity) had p values < 1 × 10−7, and GWAS for the other ethnicity was not significant. For each locus, the following are reported: locus coordinates, phenotype, and the two studies (study names as reported by the COVID-19 HGI).
(A–F) Two cases where only the multi-ethnic studies have p values passing the 1 × 10−7 threshold, suggesting that these two GWAS signals are likely associated with non-European populations. (A–C) 8:124836564-125836564 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_23andme and COVID19_HGI_B2_ALL_leave_23andme; (D–F) 6:40997035-42005196 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme.
(G–L) Three cases where European-only and multi-ethnic studies share the same lead variant, but only the European-only passes the threshold, suggesting that although the multi-ethnic studies include more individuals, they are less powered to detect the COVID-19 association. Therefore, these signals are likely associated with Europeans, rather than other ethnicities. (G–I) 7:54147894-55147894 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme; (J–L) 7:107107902-108107902 locus; A2 phenotype (very severe respiratory confirmed COVID-19 versus population): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_A2_ALL_leave_UKBB_23andme.
(M–R) Two loci (1:154605882-155673527 and 3:196626406-197627009) for which the multi-ethnic study passes the threshold, but the European-only GWAS is barely below the threshold (p = 2.6 × 10−7 and p = 2.3 × 10−7, respectively) suggesting that they are underpowered; therefore, these are likely false negative associations for the European-only, and the locus is not associated with ethnicity. (M–O) 1:154605882-155673527 locus; B2 phenotype (hospitalized COVID-19 patients versus population): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme and COVID19_HGI_B2_ALL_leave_UKBB_23andme; (P–R) 3:196626406-197627009 locus; A2 phenotype (very severe respiratory confirmed COVID-19 versus population): COVID19_HGI_A2_ALL_eur_leave_23andme and COVID19_HGI_A2_ALL_leave_23andme.